Article contents
Double trouble: untangling mixed sequence signals in bird samples with avian haemosporidian co-infections
Published online by Cambridge University Press: 04 March 2022
Abstract
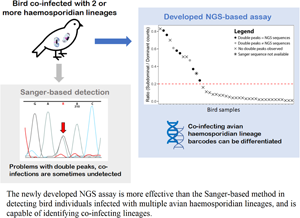
Blood parasites comprise some of the most prevalent pathogens in nature, and their detection and identification are major objectives in varied fields such as ecology and biomedicine. Two approaches were compared, one based on Sanger sequencing and the other next-generation sequencing (NGS) based, in terms of their performance in detecting avian blood parasites across tropical Southeast Asian birds. Across a panel of 528 bird individuals, 43 birds were ascertained to be infected with avian haemosporidians using a polymerase chain reaction-based detection method. Among these samples, NGS-based barcoding confirmed co-infections by multiple blood parasites in all eight cases where Sanger sequencing produced double peaks. Importantly however, the NGS-based method produced another five diagnoses of co-infections (62.5%) in which Sanger-based barcoding remained equivocal. In contrast to Sanger sequencing, the NGS-based method was able to identify co-infecting haemosporidian lineages via their barcodes. The accuracy of avian haemosporidian lineage identification was not compromised by the shorter length of NGS sequences, with ~94% of NGS barcodes producing matches identical to those of the Sanger barcodes. The application of NGS-based barcoding methods promises to enhance parasite identification and reduce erroneous inferences based on artefacts.
- Type
- Research Article
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press
References
- 3
- Cited by



