Introduction
Until recently, a large variety of lichens were accommodated in the genus Bacidia in the broad sense. Zahlbruckner (Reference Zahlbruckner, Engler and Prantl1905) used the genus name for crustose lichens with a chlorococcoid photobiont, biatorine apothecia, and ascospores with three or more transverse septa. This circumscription was apparently unnatural and included numerous taxa that are not closely related to the type species, Bacidia rosella (Pers.) De Not. (Gerasimova & Ekman Reference Gerasimova and Ekman2017). In recent phylogenetic studies, several species that have been considered within Bacidia for a long time have been transferred to other genera such as Bellicidia Kistenich et al., Bibbya J. H. Willis, Scutula Tul. and Toniniopsis Frey (Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018). Moreover, phylogenetic results showed that Toninia A. Massal. was paraphyletic and Bacidia subincompta (Nyl.) Arnold, Toninia aromatica (Sm.) A. Massal. and T. coelestina (Anzi) Vězda grouped with Toniniopsis illudens (Nyl.) Kistenich et al. (formerly Bacidia illudens (Nyl.) H. Olivier). Toniniopsis is morphologically similar to Toninia but differs in the stronger pigmentation of the exciple and the presence of a blue-green pigment in the hymenium and sometimes also in the proper exciple.
A widespread species of the genus, T. subincompta (Nyl.) Kistenich et al., is a corticolous species, occurring in Europe, Asia, Macaronesia, Africa and North America. It occurs in a range of woodlands from sea level to an altitude of c. 3000 m (Ekman Reference Ekman1996; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). It is also used as an indicator for old-growth forest and is included in indices for determining the conservation importance of woodland areas (Rücker & Wittmann Reference Rücker and Wittmann1995; Rose & Coppins Reference Rose, Coppins, Nimis, Scheidegger and Wolseley2002; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009; Brackel Reference Brackel2019). Toniniopsis subincompta can generally be recognized by its thinly granular thallus, black apothecia, green epithecium, red-brown hypothecium, and bacilliform ascospores (Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). However, it can vary in spore shape, including both bacilliform and acicular spores, and in thallus structure, ranging from either discontinuous (discrete granules or low convex areoles) to continuous (without cracks or ±rimose, wrinkled, warted, tuberculate or distinctly granular) (Ekman (Reference Ekman1996) as Bacidia subincompta). Further detailed examination has revealed differences in coloration of the apothecia (including a pale albino morph (Gilbert Reference Gilbert1996)), hypothecium and exciple, in addition to the thallus and ascospore variation.
The aim of this paper is to clarify and re-evaluate the taxonomy of the morphologically variable T. subincompta s. lat. using different approaches based on material collected from the mountain regions of Allgäu (Germany) and the North Caucasus (Russia), where T. subincompta is well represented at different elevations and on various substrata. Here we present a three-locus phylogeny of T. subincompta s. lat. and assess the species morphology based on an enlarged taxon sampling and detailed observation of type specimens. Furthermore, we applied several species delimitation programs in order to test for congruence of species differentiation based on different analyses of the molecular data.
Material and Methods
We included specimens from Germany (Allgäu mountain region; eight specimens), Russia (the North Caucasus as well as the Altay, Kaliningrad and Murmansk regions; ten specimens) and Estonia (Valgamaa and Harjumaa; two specimens) collected at different elevations and bark substrata, in the period 2013–2018 (Table 1). To test if dark and albino morphs belong to the same species, we selected specimens of both morph types from the bark of the same tree (JG145 and JG146). Voucher specimens are deposited in the herbaria of the Botanische Staatssammlung München (M), University of Tartu (TU) and Komarov Botanical Institute (LE). The herbarium collection of T. subincompta s. lat. (55 specimens) stored in the Botanical Collection of Munich (M) was also studied.
Table 1. Specimen information and DNA codes for Toninia and Toniniopsis samples used in this study, with their respective GenBank Accession numbers. New sequences are in bold.

Morphology
Microscopic observations were made using a Zeiss Axioplan light microscope (Oberkochen, Germany) equipped with differential interference contrast (DIC). Cross-sections of apothecia were made on a Leica Jung Histoslide 2000 Mikrotom (Heidelberg, Germany), with a thickness of 8–10 μm. Micrographs of cross-sections were taken on a Zeiss Axioplan with an attached AxioCam 512 Color camera and processed with the Zeiss ZEN 2.3. (blue edition) image program. Macrographs of external characters were taken on a Leica Z6 Apo microscope (with a ×2.0 Planapo lens; Leica, Germany) with a Sony Alpha 6400 camera (Sony, Japan) attached and equipped with a StackShot Macro Rail (Cognisys, USA). A single image was prepared from 30–40 serial images using Helicon Focus v.7 software (Helicon, USA).
Measurements are given as (min–) average ± SD (–max) (SD = standard deviation, n 1 = number of all observations, n 2 = number of specimens observed). We provide a detailed description of specimens using standard microscopic techniques following Coppins & Aptroot (Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009) and the subdivision scheme of the proper exciple according to Ekman (Reference Ekman1996), differentiating the following structures: the rim, lateral part and medullary part. To delimit the species lineages, we discuss the following diagnostic characters in detail: 1) thallus structure; 2) colour of disc and margin of apothecium; 3) colour of hypothecium; 4) colour and structure of exciple; 5) shape and size of ascospores. Pigment characterizations follow Meyer & Printzen (Reference Meyer and Printzen2000).
DNA extraction, PCR amplification and DNA sequencing
DNA extraction was carried out using the Stratec Invisorb Spin Plant Mini Kit (Stratec Molecular GmbH, Berlin, Germany) following the manufacturer's instructions. Five to eight apothecia were used from fresh material not older than 5 years and thallus fragments were removed in order to minimize the risk of contamination by, for example, lichenicolous fungi. PCR amplification, purification and sequencing were performed as described in Gerasimova et al. (Reference Gerasimova, Ezhkin and Beck2018). Cycling conditions included initial denaturation at 95 °C for 2 min, 5 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, 33 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, and a final extension step at 72 °C for 7 min. In those cases when the PCR product was not sufficient, a second PCR with a reduced number of cycles was conducted: denaturation at 95 °C for 2 min, 5 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, 22 cycles of 95 °C for 40 s, 54 °C for 60 s and 72 °C for 90 s, with a final extension step at 72 °C for 7 min. We used the primers ITS1F (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990) and ITS4m (Beck & Mayr Reference Beck and Mayr2012), mtSSU1 and mtSSU3R (Zoller et al. Reference Zoller, Scheidegger and Sperisen1999), and fRPB2-5F and fRPB2-7cR (Liu et al. Reference Liu, Whelen and Hall1999).
Alignment and phylogenetic analyses
In addition to 57 (18 nrITS, 19 mtSSU and 20 RPB2) newly obtained sequences of Toniniopsis subincompta s. lat., we included sequences of Toniniopsis and Toninia from GenBank in our alignment when at least two loci were available (published in Ekman Reference Ekman2001; Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018). In an additional analysis, we added to our alignment six further nrITS sequences of Toniniopsis subincompta from GenBank, collected in Switzerland (Mark et al. Reference Mark, Cornejo, Keller, Flück and Scheidegger2016) and Norway (Ekman Reference Ekman2001; Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018), in order to examine their distribution among specimens from other localities in our phylogeny (results are discussed below, and see Supplementary Material File S1, Figs S2–S4, available online).
BLAST searches in GenBank were performed to detect and exclude accessory/lichenicolous fungi and potential contaminations. Alignments were carried out using standard settings in MUSCLE v.3.8.31 (Edgar Reference Edgar2004) as implemented in PhyDE-1 v.0.9971 and optimized manually. Positions, where a gap had to be inserted in more than 95% of the sequences, were excluded. The alignment with nrITS, mtSSU and RPB2 was subjected to Bayesian inference (BI) and maximum likelihood (RAxML and IQ-TREE) analyses for the single and concatenated datasets separately.
Bayesian inference was carried out using the Markov chain Monte Carlo method (MCMC) using MrBayes v.3.2.6 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Ronquist2012). The GTR substitution model with gamma-distributed rate variations across sites and a proportion of invariable sites was selected by jModelTest 2.1.10 v20160303 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012) based on the Akaike Information Criterion. Two parallel runs were performed (two cold chains) with a single tree saved every 10th generation for a total of 1 000 000 generations. The initial 25% was discarded as burn-in and the results are summarized as a 50% majority-rule consensus tree.
Maximum likelihood (ML) analysis was performed with RAxML v.8.2.4 following a GTRGAMMA model of molecular evolution with bipartitions drawn onto the most likely tree topology using a multiple non-parametric bootstrap (Stamatakis Reference Stamatakis2014) on the CIPRES web portal (Miller et al. Reference Miller, Pfeiffer and Schwartz2010).
Further tree reconstruction using ML analysis was performed in IQ-TREE v.1.6.12 with ultrafast bootstrap approximation (Nguyen et al. Reference Nguyen, Schmidt, von Haeseler and Minh2015; Hoang et al. Reference Hoang, Chernomor, von Haeseler, Minh and Le2017). Ultrafast bootstrap (UFBoot) was specified with 10 000 replicates, 1000 maximum iterations and 0.99 minimum correlation coefficient (Minh et al. Reference Minh, Nguyen and von Haeseler2013). Substitution models for concatenated and single-locus datasets were selected using ModelFinder (Kalyaanamoorthy et al. Reference Kalyaanamoorthy, Minh, Wong, von Haeseler and Jermiin2017). The TIM2e + I + G4 substitution model was selected for the 3-locus dataset, HKY + F + G4 for the nrITS and mtSSU datasets, and TIM2e + G4 for the RPB2 dataset.
The ML trees based on the different substitution models from the single and concatenated datasets were congruent and in accordance with the Bayesian tree topology. Therefore, only the RAxML tree for the concatenated dataset is shown, with bootstrap support values (BS), posterior probabilities (PP) and ultrafast bootstrap values (UFBoot) added above or below branches. The phylogenetic trees were visualized using FigTree v.1.4.2 (Rambaut Reference Rambaut2009). Only clades with BS ≥ 70% in RAxML, PP ≥ 0.95 in BI and UFBoot ≥ 85% in IQ-TREE were considered highly supported and are indicated in bold. The concatenated and individual gene trees obtained from RAxML, MrBayes and IQ-TREE are provided in Supplementary Material File S1 (Figs S1–S13).
Species delimitation analyses
Molecular and morphological results were complemented with three widely used species delimitation computational approaches: 1) Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. Reference Puillandre, Lambert, Brouillet and Achaz2011); 2) Poisson Tree Processes (PTP) (Zhang et al. Reference Zhang, Kapli, Pavlidis and Stamatakis2013); 3) the Generalized Mixed Yule coalescent approach (GMYC) (Pons et al. Reference Pons, Barraclough, Gómez-Zurita, Cardoso, Duran, Hazell, Kamoun, Sumlin and Vogler2006). ABGD accounts for the barcoding gap (i.e., the difference between intraspecific and interspecific genetic distances) and results can be compared to morphological, geographical or ecological data (Puillandre et al. Reference Puillandre, Lambert, Brouillet and Achaz2011). We carried out ABGD using the online server at https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html, applying defaults following the recommendation of Puillandre et al. (Reference Puillandre, Lambert, Brouillet and Achaz2011). One of the most critical parameters of the ABGD method is the prior maximum divergence of intraspecific diversity (P). We assigned the range of Pmin and Pmax to 0.001 and 0.1 (defaults) but focused on the result for P = 0.01 as previous analyses have demonstrated that the method works best for this value (Puillandre et al. Reference Puillandre, Lambert, Brouillet and Achaz2011).
PTP was carried out on the online server (https://species.h-its.org/ptp/) using 100 000 MCMC generations (as recommended for small trees with < 50 taxa), saving every 100th generation with 10% discarded as burn-in. PTP is a model for delimiting species based on a phylogenetic tree. Thus, the fully resolved, optimal tree obtained from the 3-locus concatenated data from RAxML was used as an input file, but with the outgroup excluded because this can improve delimitation results (Zhang et al. Reference Zhang, Kapli, Pavlidis and Stamatakis2013).
A close relative of PTP is the GMYC model but the latter requires an ultrametric rooted tree as input with no zero branch lengths (Fujisawa & Barraclough Reference Fujisawa and Barraclough2013). To that end, identical sequences were excluded and an ultrametric tree was generated using BEAST v.1.10.4 (https://beast.community/index.html). We used BEAUti v.1.10.4 (Drummond et al. Reference Drummond, Suchard, Xie and Rambaut2012), an interactive graphical application included in the BEAST package, to generate an xml file using the following parameters: GTR + I + G model as selected by jModelTest; estimated base frequencies; a strict clock model (assumes that all branches on the tree have the same rate of evolution); Tree Prior (Speciation: Yule Process model; best suited to study the relationships between species) with a Random starting tree option (Yule Reference Yule1925; Gernhard Reference Gernhard2008), and adjusting the priors in accordance with parameters received after model calculation (parameters and table with details on prior distributions are given in Supplementary Material File S2, available online). A run of 1 M iterations logging every 100th iteration was conducted. The convergence of the Markov chain was checked using Tracer v.1.7.1. A consensus tree was generated with TreeAnotator v.1.8.2 after discarding the initial 10% of trees as burn-in. The output file was converted into a newick file using FigTree v.1.4.2 (Rambaut Reference Rambaut2009). GMYC was executed with the gmyc function in the SPLITS package in R (v.2.10; www.cran.r-project.org), employing the single (GMYCs) threshold method following parameters in the SPLITS package manual (Supplementary Material File S2). We applied the single-threshold version of GMYC since it has been shown to outperform the multiple-threshold version (Fujisawa & Barraclough Reference Fujisawa and Barraclough2013; Talavera et al. Reference Talavera, Dincă and Vila2013; Luo et al. Reference Luo, Ling, Ho and Zhu2018).
Results
Phylogenetic analyses
Eighteen new nrITS, 19 mtSSU and 20 RPB2 sequences of Toniniopsis subincompta s. lat. were obtained (Table 1). The ML and BI analyses recovered highly concordant topologies in the phylogenetic trees from single genes separately and the concatenated dataset, with Toninia cinereovirens (Schaer.) A. Massal. and T. squalida (Ach.) A. Massal. used as outgroup (outgroup selection based on Kistenich et al. (Reference Kistenich, Timdal, Bendiksby and Ekman2018)). Our phylogenetic results using three loci depict two distinct clades in Toniniopsis subincompta s. lat. (Fig. 1; Supplementary Material File S1 (Fig. S1), available online). Both clades were recovered with high support (BS/PP/UFBoot: 99/1.0/99 and 100/1.0/100, respectively). Clade I contains a highly supported subclade including JG148, 149 and 150 (BS/BI/UFBoot: 100/1.0/100).
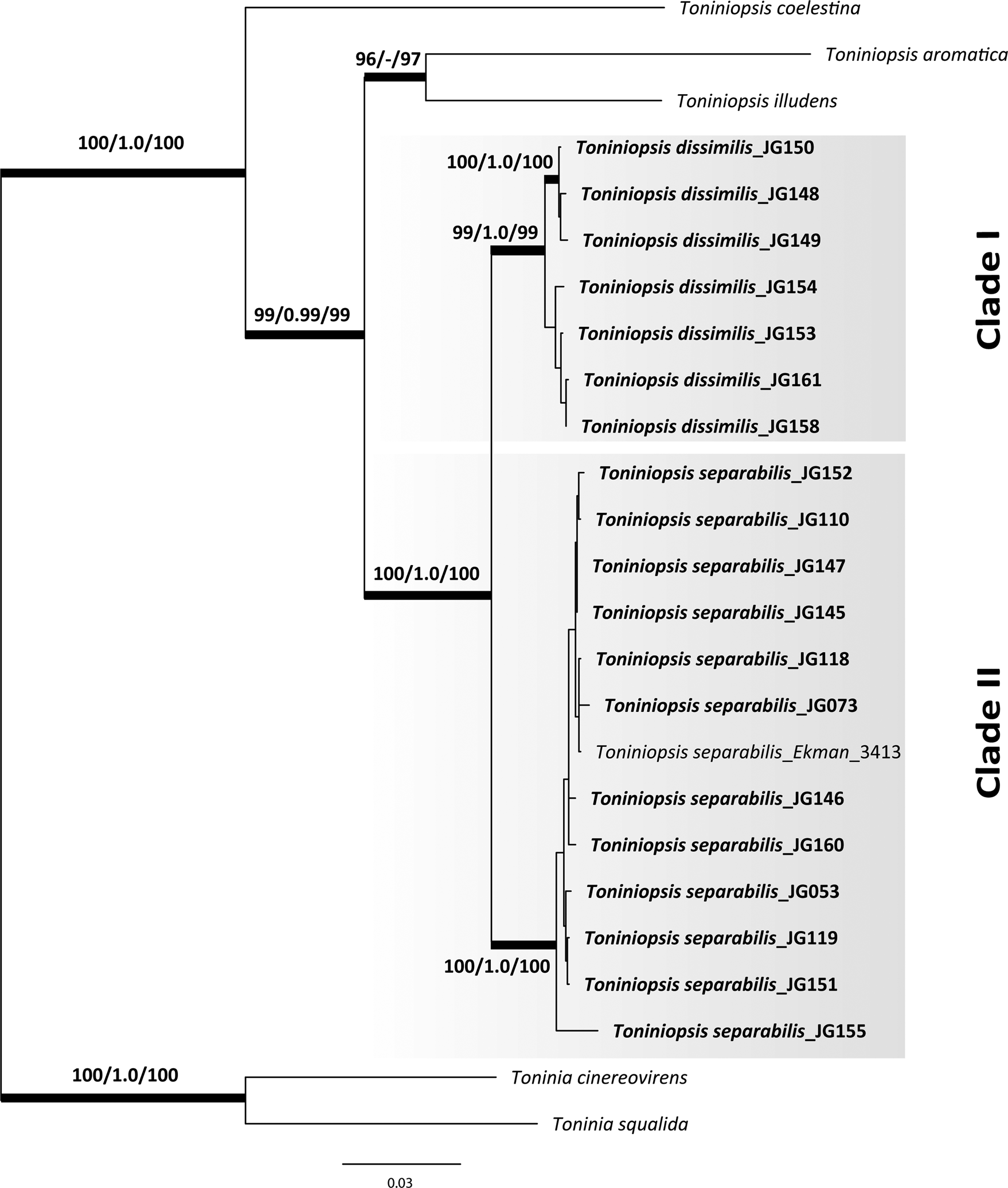
Fig. 1. Maximum likelihood (ML) tree of Toniniopsis subincompta s. lat. resulting from the RAxML analysis (Stamatakis Reference Stamatakis2014) of a concatenated 3-locus dataset (nrITS, mtSSU and RPB2). Maximum likelihood bootstrap values (BS), Bayesian posterior probabilities (PP) and ultrafast bootstrap values (UFBoot) are shown above or below branches. Branches with BS ≥ 70% in the RAxML analysis (first value), PP ≥ 0.95 (second value) and UFBoot support ≥ 85% are considered highly supported and marked in bold.
Clade I (T. dissimilis) comprises specimens from the mountain region of Allgäu and the North Caucasus collected at elevations from 1050 to 1900 m a.s.l., and Clade II (T. separabilis) contains those from the Allgäu and North Caucasus in addition to those from Estonia and other parts of Russia (Altay, Murmansk and Kaliningrad regions) collected at elevations from 4 to 1800 m a.s.l. When including six nrITS sequences of T. subincompta from GenBank collected in Switzerland (Mark et al. Reference Mark, Cornejo, Keller, Flück and Scheidegger2016) and Norway (Ekman Reference Ekman2001; Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018), it was shown that they mostly belong to Clade II, except MG838157 in Clade I, still with high support values for both clades (BS/BI/UFBoot: 99/1.0/100 and 98/1.0/95, respectively; see Figs S2–S4 in Supplementary Material File S1). Taken together, we observed no clear correlation between the clades and geography in our limited dataset. Nevertheless, individuals in Clade I have so far been collected only above 1000 m or in high latitudes (Finnmark, Norway) and thus seem to be more cold-adapted than specimens in Clade II.
Species delimitation
Molecular species delimitation programs belong to the methods within an integrative taxonomic framework and do not require a prior hypothesis of putative number of species. Species estimates for T. subincompta s. lat. using ABGD, PTP and GYMC resulted in two main species clusters in accordance with the results received from phylogenetic analyses. Thus, the first cluster included individuals from Clade I of the phylogenetic tree, and the second cluster individuals from Clade II.
Morphology
We examined the morphology and anatomy of all specimens of T. subincompta s. lat. included in our phylogeny, as well as relevant type specimens and herbarium material available in M (55 specimens). When comparing specimens from the two clades, we did not observe a strong difference in the size of the hymenium, exciple and ascospores (Table 2). The hymenium in Clade I is slightly thicker when compared to Clade II (72.5 ± 17 and 66.1 ± 11.1 μm, respectively). Ascospores in both clades are predominantly bacilliform, with 1 to 7 septa: 14–44 × 2–4 μm in Clade I, and 13–40 × 2–4 μm in Clade II. However, differences in thallus structure, as well as apothecia, exciple and hypothecium colour were observed (Table 2). Specimens in Clade I are characterized by having a predominantly wrinkled, warted to subsquamulose thallus whereas in Clade II the thallus mostly consists of single or contiguous ±loose granules, often forming short, coralloid, isidium-like bulges (Fig. 2). Apothecia in Clade I are generally grey-brown to dark brown, often with a lighter margin, whereas in Clade II apothecia are mainly darker, dark brown to black, with a margin mostly of the same colour or darker than the disc (pale brown in the albino morph). The hypothecium in Clade I is rather thick, often gradually merging into the coloration of the exciple downwards, whereas in Clade II the hypothecium is thinner and can easily be separated from the exciple below (Fig. 3). While in Clade I the rim of the exciple is mostly colourless with 2–3 layers of enlarged lumina cells (Figs 3A–E, 4A), in Clade II the rim and lateral part often contain either blue-green or brown or mixed blue-brown coloration in the upper part or along the whole margin (Fig. 3B, D & F), with mainly one layer of enlarged lumina cells (Fig. 4B). Two isolates collected from the same tree, representing the dark morph (JG145) and the albino morph (JG146) and confirmed to be the same species, nested in Clade II, but differed from other sequences by at least one nucleotide change in all three genes. In detail, the albino morph differs from the dark morph (JG145) by 4 nucleotides in nrITS, 2 nucleotides in mtSSU and one nucleotide in RPB2. Cyanotrophy, facultative or obligate association of lichens to free-living or ±lichenized cyanobacteria (Poelt & Mayrhofer Reference Poelt and Mayrhofer1988), and the occurrence of an albino morph were observed only in Clade II (see further details below). Based on the morphological and phylogenetic analyses, we conclude that there are two different species within Toniniopsis subincompta s. lat.
Table 2. Comparison of main diagnostic features between specimens from Clade I and Clade II of Toniniopsis subincompta s. lat.


Fig. 2. Detail of Toniniopsis dissimilis and T. separabilis. A, holotype of T. dissimilis (M-0290432, JG149); thallus, consisting of scattered ±rounded or flattened or subsquamulose granules, and dark apothecia with light brown margin. B, T. separabilis (LE L-15299, JG160); thallus thin, consisting of ±rounded, loose granules. C, T. dissimilis (LE L-15293, JG153); thallus thick, warted to wrinkled. D, T. separabilis (M-0182613, JG073); thallus thick, wrinkled, forming isidium-like bulges with albino morph apothecia. Scales: A–D = 0.5 mm. In colour online.

Fig. 3. Transverse sections of apothecia of Toniniopsis dissimilis (A, C, E) and T. separabilis (B, D, F). A, holotype of T. dissimilis (M-0290432, JG149). B, lectotype of Lecidea separabilis (H-NYL 17424). C, colourless rim of exciple; lateral part brown, mainly attached to hymenium, partly with blue tinge near hymenium (M-0290431, JG148). D, rim and lateral part of exciple with blue coloration along the whole margin (M-0290425, JG145; dark morph). E, dark brown hypothecium, merging into coloration of exciple below (L-15293, JG153). F, dark brown hypothecium with paler exciple below; rim and lateral part of exciple with brown coloration along the whole margin (M-0290437, JG152). Scales: A–F = 200 μm. In colour online.

Fig. 4. Exciple structure. A, holotype of Toniniopsis dissimilis (M-0290432, JG149) with at least 3 layers of enlarged lumina cells. B, lectotype of Lecidea separabilis (H-NYL 17424) with paraplectenchymatic exciple structure and one layer of enlarged lumina cells. Scales: A & B = 50 μm. In colour online.
Taxonomic treatment
The name Lecidea subincompta Nyl. was introduced as nomen novum for Lecidea anomala var. atrosanguinea Schaer. (Nylander Reference Nylander1865), because the epithet atrosanguinea was blocked by Lecidea atrosanguinea (Hoffm.) Nyl. (Nylander Reference Nylander1854) which was already in use. Lecidea anomala var. atrosanguinea Schaer. was based on Schaerer's Lichenes Helvetici exs. 212, which in fact contains specimens of Bacidia incompta (Borrer ex Hook) Anzi (Ekman Reference Ekman1996). This agrees with our observations on the type material in M (M-0308484). Hence Toniniopsis subincompta, based on Lecidea subincompta, would have to be regarded as a synonym of Bellicidia incompta (Borrer) Kistenich et al. (formerly Bacidia incompta). For this reason, Ekman (Reference Ekman1996) suggested it was appropriate to select a new, conserved type specimen for Lecidea subincompta since the epithet subincompta was well known, while Bacidia separabilis (Nyl.) Arnold, the name to be used without a conserved type, had hardly ever been used. The necessity of conservation was also stated by Kistenich et al. (Reference Kistenich, Timdal, Bendiksby and Ekman2018) when the taxon was transferred to Toniniopsis, but a formal proposal has never been made. According to the research presented here, the former T. subincompta consists of two species, and therefore a proposal to conserve L. subincompta (with a conserved type) will most likely not be approved. Consequently, T. subincompta is synonymized here with Bellicidia incompta. While working on the type material of T. subincompta s. lat., we did not find a matching candidate for Clade I and thus a new species must be described.
Toniniopsis dissimilis Gerasimova & A. Beck sp. nov. [Clade I]
MycoBank No.: MB 836224
This species is similar to Toniniopsis separabilis but differs in having a smooth to rather thick, tuberculate, warted or subsquamulose thallus and dark brown hypothecium, merging into the coloration of the exciple downwards.
Type: Germany, Bavaria, Landkreis Oberallgäu, Markt Oberstdorf, track from Oytalhaus to Käseralpe, c. 500 m east of Oytalhaus, mixed forest of Picea, Fraxinus and Salix along the River Oybach, on bark of the trunk of Fraxinus, c. 1 m above the ground, J. Gerasimova & A. Beck s. n., 27 June 2018 (M-0290432—holotype; UPS—isotype).
Thallus indeterminate, thin, partly smooth to rather thick, tuberculate or warted, sometimes consisting of an appressed and continuous crust of subsquamules, granular when on twigs of mosses, consisting of single or contiguous warts or occasionally of single ±rounded granules. Warts ±flattened or forming subsquamules, adnate to the surface, often merged, forming a thick, wrinkled, irregularly shaped surface, light green, grey-green, green, brown-green, dirty grey-green to green-brown. Photobiont chlorococcoid.
Apothecia (0.2–)0.4 ± 0.15(–0.7) mm diam. (n 1 = 77, n 2 = 8), young with margin raised above the disc, mature ±flat, later becoming convex. Disc grey-brown, dark brown to black, sometimes with mixed colour. Margin of the same colour as or paler than disc, light brown, grey, brown-grey, grey-brown to dark brown. Hymenium (51–)72.5 ± 17(–122.5) μm thick (n 1 = 35, n 2 = 8), colourless, without crystals. Epithecium greyish green or greyish blue, green-blue, partly almost colourless or brown. Hypothecium brown to dark brown, rather thick, downwards gradually merging into the coloration of the exciple. Exciple (30–)63 ± 19.3(–115) μm wide (n 1 = 34, n 2 = 8), sometimes with minute crystals, not dissolving in K. Rim colourless, without or with greyish blue or blue tinge near the hymenium or along the whole margin, with 2–3 layers of cells with enlarged lumina (2–)4.2 ± 1.2(–7) μm wide (n 1 = 40, n 2 = 8), and (5–)8.3 ± 2.9(–24) μm long (n 1 = 40, n 2 = 8; maximum value with 24 μm was observed only once in JG149). Lateral part colourless or pale brown to dark brown, often coloured mainly when attached to hymenium. Medullary part pale brown, brown to almost colourless downwards, paler than hypothecium. Paraphyses simple or septate, (1.5–)1.8 ± 0.4(–3.5) μm wide (n 1 = 42, n 2 = 8); apices ±clavate or not at all swollen, sometimes bifurcate, (1.5–)2.5 ± 0.7(–4) μm wide (n 1 = 42, n 2 = 8), colourless or with dark blue diffuse internal pigmentation. Ascospores bacilliform, (14–)26.7 ± 4.2(–44) μm long (n 1 = 245, n 2 = 8), and (2–)2.9 ± 0.4(–4) μm wide (n 1 = 245, n 2 = 8), with (1–)4 ± 2(–7) septa (n 1 = 245, n 2 = 8).
Chemistry
Hypothecium K+ intense brown to purplish; exciple K+ purplish brown.
Pigments
Bagliettoa-green in epithecium and in uppermost part and rim of exciple; Laurocerasi-brown in hypothecium and lateral part of exciple.
Etymology
Similar to T. separabilis but differs in some characters.
Additional specimens examined
Turkey: Prov. Rize: valley very near to the village of Ayder, 30 vii 1997, V. John (M-0308422). Prov. Bursa: Uludag, 6 ix 1976, K. Kalb & G. Ploebst (M-0308424).—Germany: Bayerische Alpen: am Taubensee ober Wessen, ix 1876, F. Arnold (M-0308441); ibid., ix 1871, F. Arnold (M-0308442); Aschau, ix 1875, F. Arnold (M-0308443).—Sweden: Södermanland: Björkvik, 27 vi 1908, G. O. A. Malme (M-0308448).—Norway: Troms: Tromsø, 1868, Th. M. Fries (M-0308451).—Finland: [South Häme]: Luhanka, 1873, E. Lang (M-0308456).—Slovakia: Malá Fatra: Velký Stoh, c. 1300 m, 17 vi 1965, I. Pišút & A. Vězda (M-0308458).—Austria: Steiermark: Niedere Tauern, Wölzer Tauern, 1500 m, 15 vii 1990, J. Hafellner (M-0308468). Vorarlberg: Rätikon, Gargellen, 1300 m, 6 ix 1989, R. Türk (M-0308469). Tirol: Matrei, F. Arnold (M-0308476, M-0308477).
Toniniopsis separabilis (Nyl.) Gerasimova & A. Beck comb. nov. [Clade II]
MycoBank No.: MB 836225
Lecidea separabilis Nyl., Flora (Jena) 48(10), 147 (1865).—Bacidia atrosanguinea f. separabilis (Nyl.) Arnold, Flora (Jena) 67(30), 582 (1884); type: [Finland, South Häme], Hollola, 1863, J. P. Norrlin s. n. (H-NYL 17424—lectotype! selected by Ekman (Reference Ekman1996: 105)).
Lecidea hegetschweileri Hepp [nom. nud.] in Hepp's Systematische Sammlung, 212 (1852).—Biatora atrosanguinea [unranked] hegetschweileri Hepp, Die Flechten Europas 1 (1853).—Bacidia hegetschweileri (Hepp) Vain. [non auct.], Acta Soc. Fauna Flora Fenn. 53(1), 215 (1922); type: [Switzerland], Zürich, An der Rinde alter Eichen, J. A. P. Hepp s. n. [Hepp: Flecht. Europ., 23] (BM00002217—lectotype! selected by Ekman (Reference Ekman1996: 105); M-0190116, M-0154468—isolectotypes!).
Secoliga atrosanguinea var. affinis Stizenb., Nova Acta Acad. Leopoldin.-Carolin. 30(3), 18 (1863) [non Bacidia atrosanguinea (Sch.) var. affinis Zw. in Arnold: Lich. exs. 505].—Bacidia affinis (Stizenb.) Vain., Acta Soc. Fauna Flora Fenn. 53(1), 154 (1922); type: [Germany, Baden-Württemberg], Heidelberg, in der Nähe des Kohlhofs, an Popul. tremula, sehr selten, 1858, Zwackh-Holzhausen s. n. [Lichenes exs. 336B] (H-NYL 17397—lectotype! selected here, MBT394654).
Lecidea bacillifera f. melanotica Nyl., Flora (Regensburg) 50, 373 (1867).—Bacidia subincompta var. melanotica (Nyl.) H. Magn., Förteckning över Skandinaviens växter 4, 40 (1936); type: Finland, [South Häme], Evois (= Evo), ad corticem populi, 1866, J. P. Norrlin s. n. (H-NYL 17205—lectotype! selected here, MBT394655; H-NYL 17362—isolectotype!).
Bacidia vegeta Vain., Acta Soc. Fauna Flora Fenn. 53(1), 153 (1922); type: [Finland, South Häme], Mustiala, 1868, A. Kullhem s. n. (H8000177—lectotype! annotated by S. Ekman (2012), selected here, MBT394656).
(Figs 2B & D, 3B, D & F, 4B, 5)

Fig. 5. Cyanotrophic association in lectotype of Lecidea bacillifera f. melanotica (A, B & E) and T. separabilis (C, D & F). A, thallus with cyanobacterium (dark patches) in tight association. B, composition of the thallus seen under the microscope. C, cross-section of apothecium with associated cyanobacterium cells (M-0289891, JG110). D, Nostoc sp. (M-0289891, JG110). E, Gloeocapsa sp. F, Scytonema sp. (M-0290436, JG151). Scales: A = 1 mm; B = 100 μm; C = 50 μm; D–F = 15 μm. In colour online.
Thallus indeterminate, thin, partly smooth, inconspicuous, discrete to rather thick, continuous, warted, thinly to coarsely granular, consisting of single or contiguous ±loose granules, especially rich in cracks of the bark, often forming short, coralloid, isidium-like bulges. Granules ±roundish or flattened or forming patches of irregularly shaped subsquamules, green, green-grey to green-brown, turning completely brown in the herbarium. Photobiont chlorococcoid, sometimes additionally associating with one or several free-living cyanobacteria: 1) Nostoc sp., 2) Gloeocapsa sp. and 3) Scytonema sp. (cyanotrophic association).
Apothecia (0.2–)0.4 ± 0.1(–0.8) mm diam. (n 1 = 88, n 2 = 10), young ±flat or with margin slightly above the disc, mature ±flat, later becoming convex and irregularly shaped. Disc grey-brown, dark brown to black or pale orange to orange-brown in albino morphs. Margin of the same colour as or darker than the disc, light brown, brown to dark brown to black. Hymenium (49–)66.1 ± 11.1(–98) μm thick (n 1 = 43, n 2 = 10), colourless, without crystals. Epithecium brown, greyish blue to dark blue-green or sometimes almost colourless (albino morph). Hypothecium thin, brown to dark brown, sometimes with green tinge, or straw-coloured, almost colourless in the albino morph, can be easily separated from exciple below. Exciple (59–)63.5 ± 18(–122.5) μm wide (n 1 = 39, n 2 = 10), without or with minor crystals along exciple, not dissolving in K. Rim pale brown, dark brown or blue, often mixed blue-brown, mostly with one layer or up to two layers of enlarged lumina cells along the rim, often inconspicuous, (2.0–)3.5 ± 0.8(–6) μm wide (n 1 = 55, n 2 = 10) and (5–)7.5 ± 1.8(–13) μm long (n 1 = 55, n 2 = 10). Lateral part pale brown to brown mainly attached to hymenium, often with blue, brown or mixed blue-brown colour near hymenium or along the whole margin, or almost colourless or pale straw-coloured (albino morph). Medullary part under hypothecium light brown to almost colourless which downwards can eventually change to dark brown or blue. Paraphyses simple, (1–)1.8 ± 0.5(–3.5) μm wide (n 1 = 60, n 2 = 10); apices ±clavate or not at all swollen, sometimes bifurcate, (1.5–)3 ± 1(–5.0) μm wide (n 1 = 60, n 2 = 10), colourless or with dark greyish blue to dark blue-green diffuse internal pigmentation. Ascospores bacilliform or acicular, (13–)25.2 ± 4.2(–40) μm long (n 1 = 273, n 2 = 10) and (2–)2.7 ±0.42(–4) μm wide (n 1 = 273, n 2 = 10), with (1–)4 ± 2(–7) septa (n 1 = 273, n 2 = 10).
Chemistry
Hypothecium K+ intense brown; brown parts of exciple K+ purplish brown.
Pigments
Bagliettoa-green in epithecium and rim of exciple; Laurocerasi-brown in hypothecium and lateral part of exciple; sometimes a mixture of Bagliettoa-green and Laurocerasi-brown in exciple. Rubella-orange in epithecium, hypothecium and exciple (in the case of the albino morph).
Remarks
According to the International Code of Nomenclature, for any taxon below the rank of genus, the correct name is the combination of the final epithet of the earliest legitimate name of the taxon at the same rank (Art. 11.4, Turland et al. Reference Turland, Wiersema, Barrie, Greuter, Hawksworth, Herendeen, Knapp, Kusber, Li and Marhold2018). Therefore, Lecidea separabilis has priority at species rank from 1865, whereas the epithets hegetschweileri and affinis were validly published at the species rank only in 1922 (Vainio Reference Vainio1922). The complex nomenclature regarding the name Bacidia hegetschweileri has already been described in detail by Ekman (Reference Ekman1996) so we do not reiterate this information here.
Additional specimens examined
Russia: Republic of Adygea: Maykop district, 1900 m, I. Urbanavichene & G. Urbanavichus (LE L-15296); ibid., 1460 m, I. Urbanavichene & G. Urbanavichus (LE L-15301). Kabardino-Balkaria: Elbrus National Park, 1880 m, I. Urbanavichene & G. Urbanavichus (LE L-15302).—Canada: Graham Island: Mt Raymond, c. 760 m, 1988, I. M. Brodo 26653B (M-0308421).—Turkey: Prov. Kastamonu: Ilgaz Dağlari, 1680 m, 11 viii 1976, K. Kalb & G. Ploebst (M-0308423); ibid., 1680 m, 31 viii 1976, K. Kalb (M-0308425).—Germany: Landkreis Oberallgäu: Markt Oberstdorf, 850 m, 27 vi 2018, J. Gerasimova & A. Beck (M-0290429); ibid., 1050 m, 27 vi 2018, J. Gerasimova & A. Beck (M-0290434); ibid., 950 m, 29 vi 2018, J. Gerasimova & A. Beck (M-0290439; M-0290440). Landkreis Berchtesgadener Land: Königssee, 605 m, 1984, R. Türk & H. Wunder 2961 (M-0308431); ibid., 630 m, 1998, R. Türk 25704 (M-0308426); Obersalzberg, 955 m, 2011, R. Cezanne & M. Eichler 8371 (M-0308428); Ramsau, 775 m, 1988, R. Türk & H. Wunder 5988 (M-0308429); Hochbahnweg, 1050 m, 1989, R. Türk & H. Wunder 6257 (M-0308430). Landkreis Freyung-Grafenau: National Park Bayerischer Wald, 1100–1180 m, 13 x 1999, Ch. Printzen (M-0308427). Landkreis Weissenburg-Gunzenhausen: Weissenburg, 20 v 1866, F. Arnold (M-0308432). Landkreis München: zwischen Geiselgasteig und Wörnbrunn, im Grünwalder Park, 3 x 1888, F. Arnold (M-0308434); zwischen Obersendling und Hesselohe, vi 1888, F. Arnold (M-0308436). Landkreis Starnberg: westlich von Gauting, 23 iii 1896, F. Arnold (M-0308435). Landkreis Coburg: Tal W Hohe Schwemge, NW Oberwohlsbach, 1989, L. Meinunger 17353 (M-0308437). Landkreis Miesbach: Alpbach, Tegernsee, 1989, L. Meinunger 17135 (M-0308438). Landkreis Eichstätt: Eichstätt, 1858, F. Arnold (M-0308440); ibid., v 1864, F. Arnold (M-0308444); ibid., vii 1869, F. Arnold (M-0308445). Thüringen: Kreis Hildburghausen, 1989, L. Meinunger 17004 (M-0308446).—Austria: Salzburg: Brunnfeld between the villages of Pichl-Auhof and Ort, 605 m, 5 iv 2008, R. Türk & S. Pfleger (M-0308465); Hohe Tauern, Kapruner Tal, 970–1270 m, 27 x 1989, R. Türk (M-0308466). Kärnten: Rennweg N von Pörtschach, 600 m, 14 ii 1990, R. Türk (M-0308467). Vorarlberg: Lechtaler-Alpen, 1300 m, 10 vii 1986, H. Mayerhofer, G. Grabherr & R. Türk (M-0308470). Steiermark: Oberfahrenbach, 400 m, 19 iii 1995, J. Poelt (M-0308471). Tirol: Wildpark Wildbichl, vii 1895, Schnabl (M-0308439); Matrei, vii 1869, F. Arnold (M-0308472, M-0308473); ibid., viii 1872, F. Arnold (M-0308478).—Sweden: Värmland: Ekshärad, 11 vii 1961, S. W. Sundell (M-0308447). Uppland: Värmdön, 28 vii 1908, G. O. A. Malme (M-0308449). Jämtland: Enafors, 26 vii 1910, G. O. A. Malme (M-0308450).—Norway: Troms: Tromsø, 1868, Th. M. Fries (M-0308452).—Finland: [North Häme]: Saarijärvi, Saarikylä, 6 viii 1947, A. Koskinen (M-0308453); Mahlu, Syväoja, 5 vii 1948, A. Koskinen (M-0308454); Mahlu, Lylymäki, 3 vi 1943, A. Koskinen (M-0308455). South Häme: Evo, 1874, J. P. Norrlin (M-0308457).—Slovakia: Žilina Region: Králova, Királyhegy hola, Teplička, H. Lojca (M-0308460).—Czech Republic: Moravia: Telč, ‘Roštýnská obora’, 550 m, 26 vi 1965, A. Vězda (M-0308459); Carpati, prope pagum Ostravice, 700 m, 24 viii 1964, A. Vězda (M-0308464).—Romania: Transylvania: ad ramulos abietum in regione Aragyes infra alpem Retyezát, c. 1500 m, 23 viii 1873, H. Lojka (M-0308461); ibid., 9 viii 1873, H. Lojka (M-0308462); prope Thermas Herculis in Banatu, 16 ix 1872, H. Lojka (M-0308463).
Discussion
Our results clearly demonstrate the presence of two distinct species within the former Toniniopsis subincompta. The results obtained from phylogenetic and morphological analyses were congruent with species estimates for T. subincompta s. lat. using ABGD, PTP and GYMC. Based on the material examined, mainly from Eurasia, T. separabilis is the more common taxon (76.4%) and has the wider distribution range (Canada, Norway, Sweden, Estonia, Germany, Austria, Czech Republic, Slovakia, Romania, Turkey and Russia). Toniniopsis dissimilis is less frequent (23.6%) and specimens were examined from Norway, Sweden, Finland, Germany, Slovakia, Austria, Turkey and Russia. The latter species seems to be more cold-adapted as we have only seen specimens collected above 1000 m or in high latitudes.
According to the literature, the distribution of T. subincompta s. lat. extends to Macaronesia, Africa and North America (Ekman Reference Ekman1996; Llop Reference Llop2007; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). Based on observations of specimens from North America by Ekman (Reference Ekman1996), T. subincompta has the same variation in thallus structure as we observed in specimens from Eurasia. Apothecia are mainly dark, purple-brown to black, and the exciple rim has a single layer of enlarged globose lumina cells which correspond to Clade II but are smaller in size (5 × 8 μm). In some specimens, bacilliform ascospores were found in young apothecia, whereas acicular ascospores were found in old apothecia; a variation in spore shape may even occur within the same apothecium (Ekman Reference Ekman1996, loc. cit.). We found only bacilliform ascospores in all specimens except two, but looked mainly at middle-aged apothecia. The two exceptions with long acicular ascospores were JG053 (Murmansk Region) and the type specimen of Lecidea bacillifera f. melanotica (Finland). In addition, ascospores in northern American material are longer and thicker with a larger number of septa (19–64 × 1.9–6.2 μm, with 3–13 septa). Our observations correspond with those on T. subincompta from Great Britain and Spain (ascospores bacilliform, 20–36–40 × 2.3–3.5–4 μm, with 3–7 septa and 18–40 × 2–4 μm, with 3–9 septa, respectively (Llop Reference Llop2007; Coppins & Aptroot Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). A single herbarium specimen observed from Canada was determined to be T. separabilis with characters corresponding to our description. Potentially, T. subincompta s. lat. from North America represents a separate entity to those from Eurasia. However, additional morphological and phylogenetic investigations are required.
In T. subincompta s. lat., an albino morph had already been mentioned by Coppins & Aptroot (Reference Coppins, Aptroot, Smith, Aptroot, Coppins, Fletcher, Gilbert, James and Wolseley2009). In order to test if dark and albino morphs belong to the same species, we sequenced both, a dark and an albino morph from bark of the same tree. According to our results, the two isolates belong to the same species but differ from each other in three genes. In fact, none of the sequences of dark morphs were identical to the albino morph, while identical sequences of dark morphs did occur. Further studies are required to find out if the albino morphs form a separate lineage or are formed independently by spontaneous mutation.
Cyanotrophy is defined as facultative or obligate association of lichens to free-living or ±lichenized cyanobacteria (Poelt & Mayrhofer Reference Poelt and Mayrhofer1988). We observed the strongest association with cyanobacteria in one of the type specimens, Lecidea bacillifera f. melanotica (Gloeocapsa sp. and Nostoc sp. (Fig. 5A, B & E)), which was assigned to T. separabilis in this study based on morphological observation. The strong cyanotrophic association appears to lead to a modification of the melanotica thallus, represented by black-coloured patches. Similarly, a strong connection was observed in two further herbarium specimens collected in Sweden (M-0308447 and M-0308450), which were associated with Nostoc sp. The most diverse composition of cyanobacteria was observed in JG151 (Oberallgäu), with species from three different genera: Gloeocapsa, Nostoc and Scytonema. Juvenile stages of some green-algal lichens may establish loose cyanotrophic associations with free-living cyanobacteria and/or cyanolichens (Rikkinen Reference Rikkinen2003). Such associations in Ramalinaceae are known in Thalloidima A. Massal., most species of which are parasitic on cyanolichens when young or remain parasitic (Kistenich et al. Reference Kistenich, Timdal, Bendiksby and Ekman2018). However, the specimens we analyzed were clearly associated with free-living cyanobacteria and were not cyanolichens. Previously, complementary associations with cyanobacteria in crustose lichens have mainly been described from saxicolous species, probably due to low nutrient levels in the substratum (Poelt & Mayrhofer Reference Poelt and Mayrhofer1988). However, all specimens with cyanobacteria of T. separabilis were collected on tree bark (Picea sp., Populus sp., Salix sp., Thuja sp. and Ulmus sp.). Cyanotrophy was observed only in T. separabilis and is considered to be facultative based on our observations that only 11 out of 68 specimens (including herbarium, type and sequenced specimens) examined were cyanotrophic (c. 16%). Albino morphs, which have been found in only four specimens of T. separabilis (c. 6%), were likewise not observed in specimens of T. dissimilis. These interesting aspects of lichen biology deserve further study.
Acknowledgements and author contribution
We thank the curators of the herbarium H (Helsinki) for the loan of type specimens and Ave Suija, Evgeny Davydov and Piret Lõhmus who kindly provided specimens for investigation and sequencing. We also thank Stefan Ekman (Uppsala, Sweden) for valuable taxonomic discussions and two anonymous reviewers and the editor for highly valuable comments and improvements. In addition, we are grateful to David Richardson (Halifax, Canada) for his corrections to the English text. JG was supported by a BAYHOST fellowship from the Bayerische Staatsministerium für Bildung und Kultus, Wissenschaft und Kunst. The fieldwork by GU was supported by a grant from the Russian Foundation for Basic Research (no. 15-29-02396). The study by IU was carried out as part of the institutional project (‘Herbarium collections of BIN RAS (history, conservation, investigation and replacement)’) of the Komarov Botanical Institute of the Russian Academy of Sciences (AAAA-A18-118022090078-2). Molecular work was supported by a grant to AB from the Bayerische Staatsministerium für Bildung und Kultus, Wissenschaft und Kunst within the ‘Barcoding Fauna Bavarica’ framework. The collecting permit issued by the Regierung von Schwaben to AB is gratefully acknowledged. AB and JG designed the study and wrote the paper; JG performed the experiments; GU and IU provided the material collected in the Caucasus and Murmansk Region, and approved the manuscript.
Author ORCIDs
Julia V. Gerasimova, 0000-0002-3212-3596; Gennady P. Urbanavichus, 0000-0003-3222-5151; Irina N. Urbanavichene, 0000-0002-5492-5215; Andreas Beck, 0000-0003-2875-4464.
Supplementary Material
To view Supplementary Material for this article, please visit https://doi.org/10.1017/S0024282921000013.












