Introduction
The McMurdo Dry Valleys in Antarctica, one of Earth's coldest dry deserts, contain numerous ice-covered lakes with unique geochemistry and microbial communities (Spigel & Priscu Reference Spigel and Priscu1998, Roberts et al. Reference Roberts, Priscu, Wolf, Lyons and Laybourn-Parry2004, Cavicchioli Reference Cavicchioli2015, Sohm et al. Reference Sohm, Niederberger, Parker, Tirindelli, Gunderson and Cary2020). The microbial genomic analysis of these lakes remains limited (Koo et al. Reference Koo, Hakim, Morrow, Crowley, Andersen and Bej2018, Dillon et al. Reference Dillon, Hawes, Jungblut, Mackey, Eisen, Doran and Sumner2020, W. Li et al. Reference Li, Dore, Steigmeyer, Cho, Kim and Liu2020). There are a number of reports on antibiotic resistance genes (ARGs) in Antarctic soil or marine bacteria (Tam et al. Reference Tam, Wong, Yong, Blamey and González2015, Wang et al. Reference Wang, Stedtfeld, Kim, Chai, Yang and Stedtfeld2016, van Goethem et al. Reference Van Goethem, Pierneef, Bezuidt, van de Peer, Cowan and Makhalanyane2018, Na et al. Reference Na, Wang, Gao, Li, Jin, Zhang and Zong2019, Antelo et al. Reference Antelo, Giménez, Azziz, Valdespino-Castillo, Falcón and Ruberto2021) but few studies of ARGs in Antarctic lake water or their mat communities (Jara et al. Reference Jara, Bello-Toledo, Domínguez, Cigarroa, Fernández and Vergara2020). Antibiotic biosynthesis and resistance are natural phenomena that predate the human discovery of antibiotics. Today, the human use of antibiotics has amplified the environmental prevalence of resistance. The prevalence of ARGs in relatively pristine habitats is of importance for human health, as it provides a baseline for assessment of human-associated ARG contamination (Allen et al. Reference Allen, Donato, Wang, Cloud-Hansen, Davies and Handelsman2010, D'Costa et al. Reference D'Costa, King, Kalan, Morar, Sung and Schwarz2011). Antimicrobial resistance is a leading cause of death worldwide (Murray et al. Reference Murray, Ikuta, Sharara, Swetschinski, Robles Aguilar and Gray2022).
We report on the taxonomic composition and ARG prevalence in the water of Lake Fryxell and Lake Bonney and in the phototrophic mat communities of Lake Fryxell (Fig. 1a). Lake Fryxell extends along the Taylor Valley, 10 km downstream of Lake Bonney and 80 km across the Ross Sea from McMurdo Station. No permanent stream connects the two lakes; their main hydrology involves glacial meltwater, and each lake has an underground brine aquifer (Mikucki et al. Reference Mikucki, Auken, Tulaczyk, Virginia, Schamper and Sørensen2015). Their food webs are entirely microscopic, with no fauna larger than nematodes. The lakes and their ecosystems are studied as models for ancient Mars (Head & Marchant Reference Head and Marchant2014) and as indicators of climate change (Hall et al. Reference Hall, Castro, Kenig and Doran2017).

Fig. 1. Lake Fryxell uplift mats. a. Lake Fryxell with permanent ice cover, weathered by katabatic winds (10 December 2014). b. Source of Mat-4 DNA. Uplift mat emerges from the ice (~40 cm across).
Lakes Fryxell and Bonney have permanent stratified layers in the water column that give rise to oligotrophic microbial communities (Roberts et al. Reference Roberts, Priscu, Wolf, Lyons and Laybourn-Parry2004, Li et al. Reference Li, Podar and Morgan-Kiss2016, Kwon et al. Reference Kwon, Kim, Takacs-Vesbach, Lee, Hong and Kim2017). The lack of turnover allows for the growth of benthic microbial mats that form flame-shaped towers several centimetres tall (Hawes et al. Reference Hawes, Sumner, Andersen, Jungblut and Mackey2013, Jungblut et al. Reference Jungblut, Hawes, Mackey, Krusor, Doran and Sumner2016). The microbial mats have been studied with primary emphasis on the role of cyanobacteria in mat morphology, such as Microcoleus and Oscillatoria (Taton et al. Reference Taton, Grubisic, Brambilla, de Wit and Wilmotte2003, Jungblut et al. Reference Jungblut, Hawes, Mackey, Krusor, Doran and Sumner2016). Metagenomes of lake water, however, reveal a high abundance of proteobacteria (Dillon et al. Reference Dillon, Hawes, Jungblut, Mackey, Eisen, Doran and Sumner2020) and protists (Bielewicz et al. Reference Bielewicz, Bell, Kong, Friedberg, Priscu and Morgan-Kiss2011, Li et al. Reference Li, Podar and Morgan-Kiss2016). The Fryxell sedimentary biofilm includes a layer of phototrophic purple bacteria beneath the cyanobacterial layer (Buffan-Dubau et al. Reference Buffan-Dubau, Pringault and de Wit2001).
The benthic cyanobacterial biofilms form ‘lift-off’ mats that reach upward, buoyed by oxygen bubbles derived from photosynthesis. Pieces break off and float upward to the under-surface of the ice cover (Parker et al. Reference Parker, Simmons, Wharton, Seaburg and Love1982, Moorehead et al. Reference Moorehead, Doran, Fountain, Lyons, McNight and Priscu1999). During winter, water freezes beneath the mat fragments, adding to the ice layer, but above the lake the valley's dry winds ablate the ice. Ice sections reveal six to eight annual layers of freezing below followed by ablation above (Parker et al. Reference Parker, Simmons, Wharton, Seaburg and Love1982), leading to exposure of the desiccated mats (Fig. 1b). Surviving several years trapped in ice, the mat material remains viable, and psychrophiles may continue growing (Boetius et al. Reference Boetius, Anesio, Deming, Mikucki and Rapp2015). When mat organisms reach the surface of ablated ice, they can blow off and enter perimeter ‘moats’ of melted ice that occur in summer, surrounding the persisting ice in the main part of the lake. The mat material carries communities that include microscopic arthropods and nematodes; thus, the mats can act as vectors for the transport of entire ecosystems (Parker et al. Reference Parker, Simmons, Wharton, Seaburg and Love1982, Brambilla et al. Reference Brambilla, Hippe, Hagelstein, Tindall and Stackebrandt2001, Dillon et al. Reference Dillon, Hawes, Jungblut, Mackey, Eisen, Doran and Sumner2020).
Molecular genetic data on McMurdo Dry Valley microbes remained limited until recently (Taton et al. Reference Taton, Grubisic, Brambilla, de Wit and Wilmotte2003, Vick-Majors et al. Reference Vick-Majors, Priscu and Amaral-Zettler2014, Kwon et al. Reference Kwon, Kim, Takacs-Vesbach, Lee, Hong and Kim2017). In December 2014 (i.e. during the summer), we obtained water samples from Fryxell and Bonney as well as desiccated mat samples emerging from the ice cover of Fryxell. For the present study, we applied tools of metagenome analysis to compare the taxonomic diversity and ARG abundance of mat and lake samples. We also sequenced the genome of a novel ecotype of Rhodoferax antarcticus from a mat-forming enrichment culture, whose phenotype differs markedly from planktonic isolates of this species (Madigan et al. Reference Madigan, Jung, Woese and Achenbach2000, Jung et al. Reference Jung, Achenbach, Karr, Takaichi and Madigan2004, Baker et al. Reference Baker, Riester, Skinner, Newell, Swingley and Madigan2017).
Our study addressed the important question of ARG prevalence in relatively pristine Antarctic lakes. A modest level of antibiotic resistance is an ancient, widespread phenomenon naturally and historically occurring in all environments (Allen et al. Reference Allen, Donato, Wang, Cloud-Hansen, Davies and Handelsman2010, D'Costa et al. Reference D'Costa, King, Kalan, Morar, Sung and Schwarz2011). Non-anthropogenic processes can select for ARGs in pristine habitats; for example, cyanobacterial blooms drive increases in bacterial ARG prevalence and diversity (Zhang et al. Reference Zhang, Zhang, Lu, Peijnenburg, Gillings and Yang2020). But inputs of human origin can add substantially to the native ARG pool as well as add additional types of ARGs (Tam et al. Reference Tam, Wong, Yong, Blamey and González2015, Jara et al. Reference Jara, Bello-Toledo, Domínguez, Cigarroa, Fernández and Vergara2020, Antelo et al. Reference Antelo, Giménez, Azziz, Valdespino-Castillo, Falcón and Ruberto2021). We surveyed our mat and water metagenomes for the presence of ARGs, referenced to the Comprehensive Antibiotic Resistance Database (CARD; Alcock et al. Reference Alcock, Raphenya, Lau, Tsang, Bouchard and Edalatmand2020).
Methods
Sample collection and culture
Microbial communities were sampled in December 2014 from two meromictic lakes of the Taylor Valley, Victoria Land, Antarctica. Lake Fryxell has a maximum depth of 20 m (Lawrence & Hendy Reference Lawrence and Hendy1985), while Lake Bonney has a depth of 40 m (Priscu & Spigel Reference Priscu and Spigel1996). Both lakes are covered by a perpetual ice layer that is ~4 m thick (Priscu Reference Priscu2018), although summer melting occurs near the shoreline.
Microbial lift-off mat samples (Mat-01–06) were collected from independent mat tufts emerging separately from the Lake Fryxell ice surface, within the GPS area of: -77.60491, 163.16315; -77.60473, 163.16290; -77.60463, 163.16405; -77.60495, 163.16495. Each sample consisted of a separate tuft of desiccated microbial mat, collected with alcohol-sterilized forceps and stored at -20°C (4 weeks) then at -80°C (indefinitely).
Lake water was sampled from the permanent chemoclines of Lake Fryxell (-77.605, 163.163, 9 m depth; samples FRY-01, -02, -03) and Lake Bonney, east lobe (-77.719, 162.283, 15 m depth; samples BON-01, -02, -03). Samples were obtained using ice holes established by the McMurdo Dry Valleys Long-Term Ecological Research (LTER) programme (Priscu Reference Priscu2021a, Reference Priscu2021b, Reference Priscu2022a, Reference Priscu2022b). The LTER geochemical data are presented in Table I. Water samples were collected in 5 l cubitainers pre-washed in 10% HCl. Each water sample was concentrated by filtration onto 47 mm Pall Supor® 450 polyethersulfone membranes (0.45 μm pore size; Pall Corporation, NY, USA).
Table I. Physical, chemical and biological parameters from lakes Fryxell and Bonney (east lobe).

Data were retrieved from the McMurdo Dry Valleys Long-Term Ecological Research (LTER) programme (Priscu Reference Priscu2021a, Reference Priscu2021b, Reference Priscu2022a, Reference Priscu2022b).
PAR = photosynthetically active radiation; SRP = signal recognition protein.
Enrichment culture for anaerobic phototrophs was performed using Harwood photosynthetic medium (PM) supplemented with 10 mM succinate (Kim & Harwood Reference Kim and Harwood1991, Rey et al. Reference Rey, Oda and Harwood2006, Fixen et al. Reference Fixen, Oda and Harwood2019). Screwcap Pyrex tubes were filled with medium and inoculated with ~0.05 g desiccated material from sample Mat-04. Sealed tubes were incubated under ~10% photosynthetically active radiation at 10°C for 5 weeks. Portions of biofilm were serially sub-cultured for 2 week periods, then frozen at -80°C. Gram staining was performed using standard methods (Remel Gram Stain Kit; ThermoFisher Scientific, MA, USA).
Testing for the growth range of pH and NaCl amendment was performed using a Rhodoferax medium modified from references (Tayeh & Madigan Reference Tayeh and Madigan1987, Madigan et al. Reference Madigan, Jung, Woese and Achenbach2000). The medium contained yeast extract (0.5 g/l), ethylenediaminetetraacetic acid (EDTA; 20 mg/l), malic acid (4 g/l), (NH4)2SO4 (1 g/l), MgSO4⋅7H2O (200 mg/l), FeSO4⋅7H2O (12 mg/l), K2HPO4 (0.9 g/l), KH2PO4 (0.6 g/l), 10 mM sodium succinate and 100 mM 3-(N-morpholino)propanesulphonic acid (MOPS) adjusted to pH 7.0 with KOH. In addition, 1 ml/l of a trace element solution was added (H3BO3 (2.8 g/l), MnSO4⋅H2O (1.6 g/l), Na2MoO4⋅2H2O (0.76 g/l), ZnSO4⋅7H2O (240 mg/l), Cu(NO3)2⋅3H2O (40 mg/l), CoCl2⋅6H2O (200 mg/l)). For pH 6.0, the MOPS buffer was replaced with 100 mM 2-(N-morpholino)ethanesulphonic acid (MES) and for pH 8.0, the buffer was 100 mM (tris(hydroxymethyl)methylamino)propanesulphonic acid (TAPS). All buffered media were adjusted for pH with KOH. Growth of the enriched culture of R. antarcticus JLS was compared with culture of the type strain R. antarcticus ANT.BR obtained from the American Type Culture Collection (ATCC 700587).
DNA isolation and sequencing
Mat DNA was extracted using the PowerBiofilm kit (MO BIO, Carlsbad, CA, USA). From each sample, ~20 mg of material was extracted, and 0.75–1.50 μg DNA (measured using a Qubit fluorometer; ThermoFisher Scientific) was sent for sequencing. From lake water filters, 300–400 ng of DNA was obtained by extraction using an MP FastDNA SPIN DNA kit (MP Biomedicals, CA, USA) (Bielewicz et al. Reference Bielewicz, Bell, Kong, Friedberg, Priscu and Morgan-Kiss2011). All metagenomic DNA was sequenced at the Department of Energy (DOE) JGI (Joint Genome Institute Community Science Program award 1936). Shotgun metagenomic library construction and sequencing were carried out at JGI using standard protocols on the Illumina (San Diego, CA, USA) HiSeq 2500 platform. The raw reads were quality filtered using JGI standard protocols, and 1.2 Tb of total sequences were obtained. Illumina reads were further processed using Trimmomatic (Bolger et al. Reference Bolger, Lohse and Usadel2014) to remove adapters and low-quality sequences. Trimmomatic counts the number of reads per FASTQ file, and it was used to determine the total reads per sample metagenome.
Cultured Rhodoferax biofilm DNA was isolated using the PowerBiofilm kit (MO BIO). Sequencing was performed by the Michigan State University Genomics Core. Libraries were prepared using the Illumina TruSeq Nano DNA Library Preparation Kit. Sequencing was performed on an Illumina MiSeq done in a 2 × 250 bp format using an Illumina 500 cycle v2 reagent cartridge. The raw reads were quality filtered and 1 Tb of total sequences was obtained.
Identification of ARGs by ShortBRED
ShortBRED (Kaminski et al. Reference Kaminski, Gibson, Franzosa, Segata, Dantas and Huttenhower2015) is a pipeline that identifies protein family sequences by generating specific peptide markers from a database of interest, then screening the markers for specificity against the UniProt universal database of protein sequences (Bateman et al. Reference Bateman, Martin, Orchard, Magrane, Agivetova and Ahmad2021). The shortbred_identify command scans a reference database for protein family-specific peptide sequences. It was used to scan UniRef (the UniProt protein reference database) for sequences of the ARGs in version 3.0.7 of the CARD (Alcock et al. Reference Alcock, Raphenya, Lau, Tsang, Bouchard and Edalatmand2020). The minimum marker length accepted was 8 amino-acid residues, and the maximum length for combined markers for a given protein was 200. Markers that matched non-specific genes in UniProt were discarded from the marker dataset.
The ShortBRED marker set was then used by the shortbred_quantify command to assign ARGs in the 150 kb reads from lake water and mat samples. From each target read, six reading frames were translated using tblastn. A marker hit required a 95% match to the target. Sample class differences for ARG abundance were tested for significance (P ≤ 0.05) using the Kruskal-Wallis test.
Taxa classification of metagenomic reads
The 150 kb sequence reads from mat and water metagenomes were assigned to taxonomic classes using the Kraken 2 classifier pipeline (Wood & Salzberg Reference Wood and Salzberg2014, Wood et al. Reference Wood, Lu and Langmead2019). Kraken associates genomic k-mers (short sequence strings) in the reads with the lowest common ancestor taxa. The read classifications were then used to calculate taxa percentages via the Bracken abundance estimator (Lu et al. Reference Lu, Breitwieser, Thielen and Salzberg2017). The k-mer length was set at 150. For the Kraken 2-Bracken pipeline, the reference database used was the 5/17/2021 Standard Collection accessed at https://benlangmead.github.io/aws-indexes/k2.
Eukaryotic taxa were further characterized using the pipeline EukDetect (Lind & Pollard Reference Lind and Pollard2021). This pipeline assigns reads to a database of 521,824 marker genes from 241 gene families out of 3713 genomes and transcriptomes of fungi, protists and invertebrates. EukDetect identified protist species such as Geminigera cryophila (van den Hoff et al. Reference Van den Hoff, Bell and Whittock2020), Mesodinium rubrum (Yih et al. Reference Yih, Hyung, Hae, Myung and Young2004), Nannochloropsis limnetica (Kong et al. Reference Kong, Ream, Priscu and Morgan-Kiss2012) and Chlamydomonas sp. ICE-L (Lizotte et al. Reference Lizotte, Sharp and Priscu1996).
For an alternative classifier, mat and water metagenomes were mapped to core taxonomic markers using the MetaPhlAn2 pipeline (Segata et al. Reference Segata, Waldron, Ballarini, Narasimhan, Jousson and Huttenhower2012, Truong et al. Reference Truong, Franzosa, Tickle, Scholz, Weingart and Pasolli2015). MetaPhlAn2 assigns short reads to taxa using a set of marker genes identified from ~17,000 microbial reference genomes, primarily bacteria and archaea.
Genome assembly by breseq
The pipeline breseq 0.35.6 (Deatherage & Barrick Reference Deatherage and Barrick2014) was used to assemble reads from two samples of the enrichment culture obtained from Mat-04. The reads were mapped to the R. antarcticus ANT.BRT (DSMZ24876) reference genome (Baker et al. Reference Baker, Riester, Skinner, Newell, Swingley and Madigan2017).
Results
Taxa abundance of mat and water samples
Metagenomes were sequenced from three microbial sources: the water columns of Lake Fryxell (FRY-01, -02, -03) and Lake Bonney (BON-01, -02, -03) and the lift-off mat from the ice surface of Lake Fryxell (Mat-01–06). A total of 4 billion reads were sequenced, with a range of between 210 and 420 million reads per metagenome. The average read length was 147 bp (SD = 26).
From the three groups of samples, we characterized the taxa abundance using the Kraken 2-Bracken pipelines. The ice-surface mat samples (Mat-01–06) showed substantial amounts of cyanobacteria, ranging from 9% to 60% of the total (Fig. 2a). The predominant orders of cyanobacteria were Oscillatoriales and Nostocales (Fig. 2b). A high abundance of Oscillatoriales and Nostocales is consistent with previous reports for Fryxell mats obtained from benthic samples (cited in the ‘Introduction’ section). For Mat-01, Mat-02, Mat-03 and Mat-06, the cyanobacterial assignments were primarily Oscillatoriales and Nostocales. Mat-04 and Mat-05, however, showed depletion of Oscillatoriales, with a predominant abundance of Betaproteobacteria. Throughout the six mat samples, other taxa with significant abundance included Actinobacteria, Bacteroidetes and Alphaproteobacteria, with smaller abundances of Planctomycetes, Bacteroidetes and Firmicutes. Thus, the ice-trapped, air-exposed community showed a composition remarkably similar to that of the benthic mat samples from which lift-off mats originate.
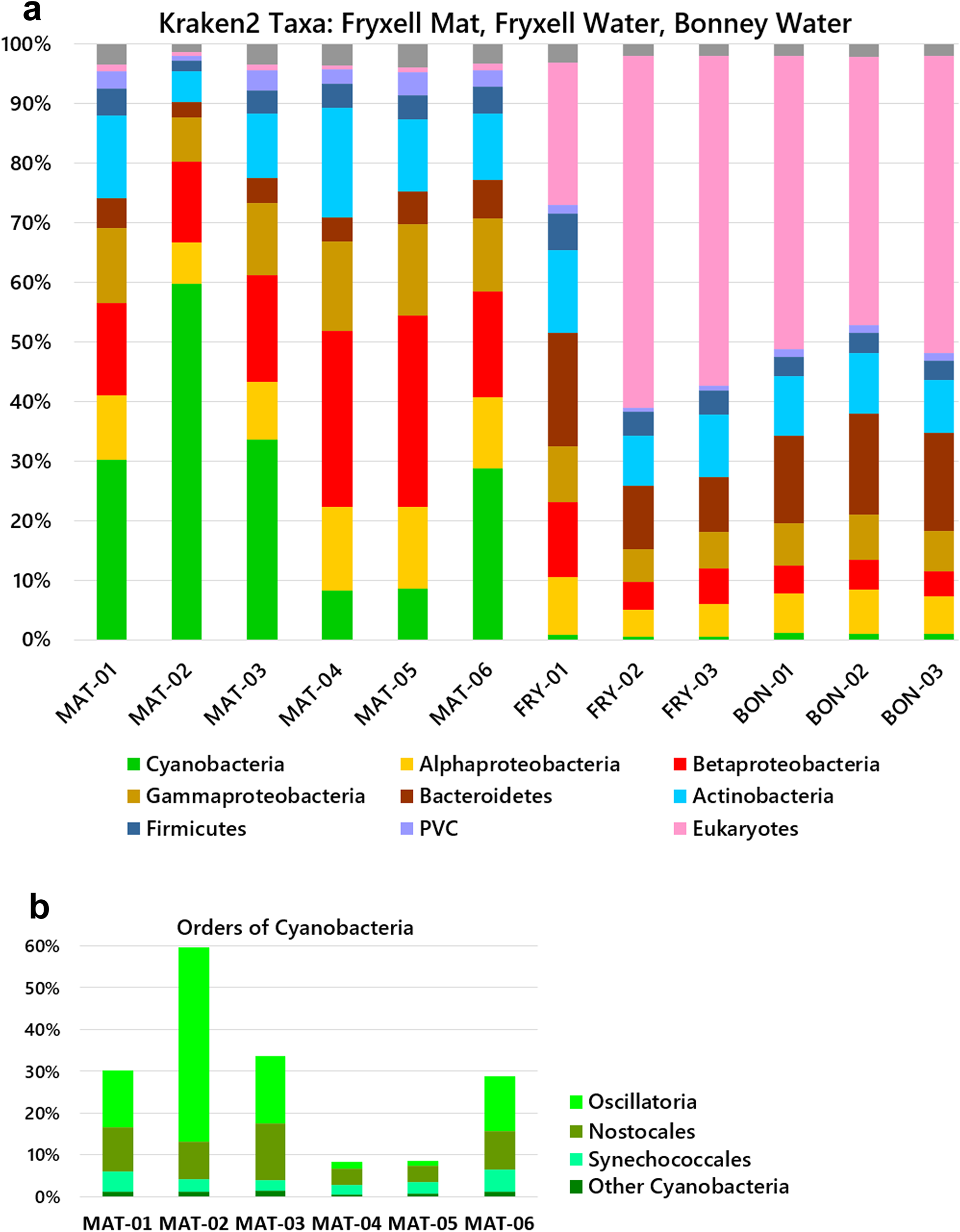
Fig. 2. Microbial community compositions of the mat and water samples. Relative abundances of phyla were determined by read alignment to marker genes using the Kraken 2-Bracken pipeline. a. Phyla and classes. b. Orders of cyanobacteria in mat samples. MAT (Lake Fryxell, mat samples); FRY (Lake Fryxell, water samples); BON (Lake Bonney, water samples). MAT samples were from the ice surface and unfiltered. The water samples were collected on a 0.45 μm filter. PVC = Planctomycetes-Verrucomicrobia-Chlamydiae.
The filtered water from both lakes Fryxell and Bonney contained abundant bacterial taxa of Actinobacteria, Bacteroides, Betaproteobacteria and Alphaproteobacteria, similarly to previous reports of Fryxell water and other glacier-associated water bodies (see the ‘Introduction’ section). The water column showed almost no cyanobacteria but a high proportion of eukaryotes, in some cases > 50% of the reads classified by Kraken 2-Bracken. By contrast, the mat samples showed virtually no detectable eukaryotic DNA. The one-sided Mann-Whitney U test confirms that water from each lake contains more eukaryotic DNA than the mat samples (P = 0.01) and that the mat samples contain more cyanobacteria than the water microbiomes of either lake (P = 0.01).
To classify the eukaryotes, we used EukDetect, a pipeline that matches short read data to marker genes from fungal, protist and invertebrate genomes (Fig. 3; full output provided in Table S1). In the lake water, EukDetect identified the protist species G. cryophila, M. rubrum, N. limnetica and Chlamydomonas sp. ICE-L. The two lakes showed similar taxa profiles, except that Chlamydomonas was found only in Lake Bonney. The mat samples had too few eukaryotic read counts to classify them.

Fig. 3. Eukaryotic taxa classified by EukDetect. Relative abundances of protist taxa were determined by read alignment to marker genes using EukDetect (Lind & Pollard Reference Lind and Pollard2021).
The validity of taxa classifier pipelines is highly dependent on their algorithm and taxa database. For comparison with the Kraken 2-Braken output, our mat and water reads were mapped to core taxonomic markers using an alternative pipeline, MetaPhlAn2 (Fig. 4). Four of the six mat samples showed mainly cyanobacteria, consistent with longstanding reports of cyanobacterial mats (see the ‘Introduction’ section). Consistent with the Kraken 2 analysis, the Mat-04 and Mat-05 samples showed major amounts of Betaproteobacteria and Alphaproteobacteria, with depletions of cyanobacteria. For the filtered water samples, the MetaPhlAn2 markers include a limited content of Eukarya, and the pipeline did not find the eukaryotic taxa predicted by Kraken 2 (Fig. 2). The filtered water from both lakes showed mainly Actinobacteria, with some Betaproteobacteria. Overall, MetaPhlAn2 appeared less effective than Kraken 2-Bracken at classifying our samples.

Fig. 4. Microbial community compositions via MetaPhlAn2. Relative abundances of class-level taxa were determined by read alignment to marker genes using MetaPhlAn2 (Segata et al. Reference Segata, Waldron, Ballarini, Narasimhan, Jousson and Huttenhower2012, Truong et al. Reference Truong, Franzosa, Tickle, Scholz, Weingart and Pasolli2015). Samples were as indicated for Fig. 2.
ARG composition characterized using ShortBRED
Gene sequences encoding various forms of antibiotic resistance, including genomic loci as well as mobile elements, are collected in the CARD database. We used the ShortBRED pipeline to identify ARGs in our samples based on a set of marker peptides matching CARD sequences. The percentage of reads from each sample that matched ARGs ranged from 0.0001% to 0.0006% (Fig. 5). The overall ARG abundances were similar amongst the three sample classes (Fryxell mat, Fryxell water, Bonney water).

Fig. 5. Antibiotic resistance gene (ARG) abundances in lake samples. Percentages of reads matched to ShortBRED markers. Total reads were counted using Trimmomatic. Bars indicate total ARG hits per sample (orange) and top 60 ranked ARG hits (blue).
The specific ARGs identified by ShortBRED were sorted by abundance across samples (Tables II & S2). Eight of the top 20 sorted ARGs showed a difference among the three sample classes (Kruskal-Wallis test, P ≤ 0.05); of these results, seven of eight are significant. Of the top-ranked ARGs, matches to CARD families BUT-1 and vancomycin resistance genes vanYA, vanTG and vanI appeared mainly in the water column. BUT-1 is a class-C beta-lactamase reported in Buttiauxella agrestis, an environmental Gammaproteobacterium found in environmental water sources. The vancomycin resistance genes occur in Enterococcus and other Gram-positives. By contrast, matches to the beta-lactamase AAC(3)-Ia occurred only in the mat samples. AAC(3)-Ia is encoded on Pseudomonas aeruginosa integrons as well as in other Proteobacteria. Other ARGs showed possible differences in prevalence between Lake Bonney and Lake Fryxell water; for example, matches to the Streptomyces-associated beta-lactamase AAC(3)-VIIa were more abundant in Bonney water, whereas vanYA matches were more abundant in Fryxell water.
Table II. Mat and water antibiotic resistance genes (ARGs) identified using ShortBRED marker peptides.

Sample class differences for ARG abundance were tested for significance (P ≤ 0.05) using the Kruskal-Wallis test. Colour-shaded rows represent sample classes for which the given ARG shows significant difference in abundance amongst the Mat, Lake Fryxell and/or Lake Bonney samples. Under the P column, yellow highlighting represents significant difference.
Overall, the three sample classes each had distinctive ARG-abundance signatures, as indicated by Kruskal-Wallis tests. Ten of the top 20 ranked ARGs were associated with Proteobacteria, which corresponds approximately with the proportion of bacterial taxa abundances predicted by Kraken 2-Bracken (Fig. 2).
Enrichment of mat-forming R. antarcticus
The relatively high proportions of Betaproteobacteria and Alphaproteobacteria in Mat-04 differed from those of other samples, and the colour of the sample material was more red than green. We cultured the various samples anaerobically (in closed tubes) at 10°C with illumination. From Mat-04, red spots of biofilm were obtained. There was no evidence of planktonic growth (cloudiness) or cell motility within the culture medium, but the organism grew as a mat, forming red blobs with a ‘red nose’ appearance. In addition, the culture extended as a red film-like mat growing along the inside of the culture tube (Fig. 6a). It was not possible to obtain isolated colonies, but serial culture of the ‘red noses’ showed consistent biofilm formation and Gram-stain morphology (Fig. 6b). This finding is consistent with Buffan-Dubau et al.'s (Reference Buffan-Dubau, Pringault and de Wit2001) report of a layer of phototrophic purple bacteria beneath the cyanobacterial layer. Ours is the first report of such a biofilm cultured from material at the ice surface rather than from its inferred origin in the sediment at the bottom of the lake.

Fig. 6. Mat-forming enrichment of Mat-04 Rhodoferax antarcticus JLS. a. Biofilm of cultured R. antarcticus within a closed tube of medium, showing a globular form of growth. The initial Mat-04 sample was sub-cultured serially three times in Harwood photosynthetic medium in horizontal closed tubes at 10°C with 30 μmol photons m-2 s-1 illumination. b. Gram stain of culture, 1000× with oil immersion. Courtesy of Emma Stuart-Bates.
The Mat-04 biofilm culture was subjected to genome analysis. DNA was sequenced and short reads were used to BLAST the National Center for Biotechnology Information (NCBI) database. The predominant hits were to the genome of R. antarcticus ANT.BRT, a Betaproteobacterium originally isolated by Madigan et al. (Reference Madigan, Jung, Woese and Achenbach2000) from a saltwater pond at Cape Royds, Ross Island, Antarctica. Kraken 2-Bracken analysis of sequenced reads agreed with this classification, assigning the samples as 95% R. antarcticus with ~1% Pseudomonas.
We used the breseq pipeline to assemble reads from two non-axenic samples (7-RN1 and 8-RN2) whose DNA sequence reads were assembled to the R. antarcticus ANT.BRT reference genome (Tables S3 & S4). For both samples, 7-RN1 and 8-RN2, 78% of reads matched the ANT.BRT reference genome. Tables S3 & S4 present ‘mutations’; that is, all of the differences from the ANT.BR reference sequence that show 100% read coverage. The two samples, 7-RN1 and 8-RN2, showed 276 and 271 sequence differences, respectively, in their main chromosomes compared with that of Baker et al.'s (Reference Baker, Riester, Skinner, Newell, Swingley and Madigan2017) reference genome (length 3,809,266 bp). Their 16S rRNA sequences showed 100% identity in breseq output. The sample genomes showed average nucleotide identity values of > 99.99% identity with the main chromosome and > 99% identity with one plasmid in the reference genome. A large part of the observed sequence differences from the reference genome were silent mutations. These observations confirm the close relatedness of our culture to the published strain.
The R. antarcticus enrichment culture (designated R. antarcticus JLS) was tested for ARG abundance using ShortBRED (Table III). More than 30 ARGs showed hits, but nearly all were associated with P. aeruginosa. By contrast, no ARGs matched those from CARD database that were associated with Rhodoferax.
Table III. Rhodoferax enrichment culture antibiotic resistance genes identified using ShortBRED.

For enrichment cultures A and B, taxa proportions were estimated using the Kraken 2-Bracken pipeline.
Characterization of the mat-forming R. antarcticus JLS
The R. antarcticus JLS biofilm was further tested for growth range in terms of pH and salinity. The biofilm was sub-cultured anaerobically with illumination in a malate-succinate Rhodoferax medium modified from that of references as described in the ‘Methods’ section (Fig. 7a–c). After 45 days, at pH 7, the bacteria formed globules as well as a red coating along the glass. At pH 6, spots of biofilm grew slowly, and at pH 8 little growth was seen. Growth was also tested for cultures buffered at pH 7 with NaCl amendment (Fig. 7d–f). The fullest growth was seen in the absence of NaCl amendment (the core medium contains ~10 mM Na+ ions). Less growth was seen with added NaCl (23 or 46 mM).

Fig. 7. pH and NaCl dependence of Rhodoferax antarcticus JLS. a.–c. Harwood photosynthetic medium containing a. 100 mM 2-(N-morpholino)ethanesulphonic acid (MES) pH 6, b. 100 mM 3-(N-morpholino)propanesulphonic acid (MOPS) pH 7 and c. 100 mM (tris(hydroxymethyl)methylamino)propanesulphonic acid (TAPS) pH 8. Cultures were incubated for 45 days. d.–f. PM medium amended with d. no NaCl added, e. 23 mM NaCl and f. 46 mM NaCl. Cultures were incubated for 45 days.
Despite the high sequence identity, the R. antarcticus JLS enrichment culture differed from Madigan et al.'s (Reference Madigan, Jung, Woese and Achenbach2000) R. antarcticus ANT.BR strain in its growth phenotype, under all conditions of pH and NaCl concentration that were tested for both strains. Unlike the motile, planktonic R. antarcticus ANT.BR, our culture showed no motility and little sign of planktonic single-celled growth. Instead, the sub-cultured material grew entirely as a biofilm, in globular spots and as a film-like growth along the interior surface of the glass tube.
The type strain ANT.BR was cultured at the same time, under all conditions. The original strain never formed a biofilm; it appeared planktonic and motile under all conditions of pH and NaCl concentration.
Discussion
Identification of ARGs from relatively pristine environments is of interest for several reasons. Long before the human introduction of high-dose antibiotics, environmental bacteria evolved multidrug pumps to efflux toxic products of their own metabolism, as well as antimicrobial substances produced by their competitors (Allen et al. Reference Allen, Donato, Wang, Cloud-Hansen, Davies and Handelsman2010, Wright Reference Wright2019). Many antibiotics possess signalling capabilities and other unknown functions. Phylogeny dates the origin of beta-lactamases to hundreds of millions of years ago. Vancomycin resistance genes are found in 30,000 year-old permafrost (D'Costa et al. Reference D'Costa, King, Kalan, Morar, Sung and Schwarz2011). But the specific kinds of ARGs found in environmental sources may differ from those prevalent in human microbiomes - those conferring resistance to the drugs we depend on for therapy (Zeng et al. Reference Zeng, Pan, Yang, Hou, Zeng and Xiong2019).
In our Taylor Valley lake metagenomes, the top-scoring ARG was BUT-1, a cephalosporinase related to sequences previously found in a clinical isolate of Buttiauxella (Fihman et al. Reference Fihman, Rottman, Benzerara, Delisle, Labia, Philippon and Arlet2002). Two other ARGs related to those of clinical origin (vanYA, vanTG) encode vancomycin resistance components in Enterococcus (Boyd et al. Reference Boyd, Du, Hizon, Kaplen, Murphy and Tyler2006, Courvalin Reference Courvalin2006). The possible finding of clinical ARGs in Antarctic water bodies is concerning. The rest of the top 20 ARGs we found appear common in environmental organisms. Eight were beta-lactamases, which are commonly found in environmental organisms but can also be readily transferred between environmental and pathogenic strains (Hooban et al. Reference Hooban, Joyce, Fitzhenry, Chique and Morris2020).
In polar regions, previous metagenomic studies reveal a range of naturally occurring ARGs. Surface soils of the Antarctic Mackay Glacier region show ARGs encoding multidrug pumps, beta-lactamases and aminoglycoside inactivators, largely associated with Gram-negative bacteria (van Goethem et al. Reference Van Goethem, Pierneef, Bezuidt, van de Peer, Cowan and Makhalanyane2018). A study of Tibetan soils showed a number of abundant ARGs, most notably encoding vancomycin resistance (B. Li et al. Reference Li, Chen, Zhang, Liu and Yan2020). Our study of ARGs from Antarctic lakes adds to this picture, showing that both water and microbial mat sources contain familiar ARGs, and probably contain others not yet discovered in Antarctic strains.
It was interesting to compare the total ARG abundance of our Antarctic lake samples with those of temperate-zone Ohio rural water bodies with moderate human inputs from a study in which samples were prepared using the same method and analysed using the same ShortBRED marker set (Murphy et al. Reference Murphy, Barich, Fennessy and Slonczewski2021). The overall ARG abundance is approximately the same for the Antarctic vs Ohio environmental sources, except for river samples obtained just downstream of a wastewater effluent pipe, where the total ARG prevalence is increased approximately five-fold. This result is consistent with a model that natural microbial communities generally harbour a small, balanced prevalence of ARGs, which gets amplified by concentrated human input. Note, however, that a database such as CARD can only represent a fraction of the actual ARGs out there; new drug resistance families and related mobility agents are continually being discovered.
The mat lift-off samples we obtained from the ice surface showed a range of DNA taxa consistent with those reported for samples obtained from benthic mats (Dillon et al. Reference Dillon, Hawes, Jungblut, Mackey, Eisen, Doran and Sumner2020). Given that our collected mat organisms had survived several years within ice, followed by air desiccation and prolonged ultraviolet radiation exposure, it is impressive how many of the Antarctic mat microbiomes retain viability with intact DNA. Even ciliated protists and invertebrate worms were obtained alive from samples cultured after months of storage at -80°C (not shown). It is probable that some of these organisms are psychrophiles that continue to grow within the ice (Boetius et al. Reference Boetius, Anesio, Deming, Mikucki and Rapp2015).
Previous studies emphasize the cyanobacterial content of lift-off mats, primarily Oscillatoriales genera such as Microcoleus, as well as Nostoc (Taton et al. Reference Taton, Grubisic, Brambilla, de Wit and Wilmotte2003, Jungblut et al. Reference Jungblut, Hawes, Mackey, Krusor, Doran and Sumner2016). While most of our lift-off samples showed an abundance of cyanobacteria, one sample yielded cultures from which the majority of reads matched R. antarcticus. The finding of mat samples enriched for R. antarcticus indicates that portions of the lower layer of the Rhodoferax mat (Buffan-Dubau et al. Reference Buffan-Dubau, Pringault and de Wit2001), along with the cyanobacterial upper layer, can break off and form lift-off patches that emerge from the ice. Our culture of R. antarcticus (R. antarcticus JLS) obtained from Lake Fryxell showed a mat-forming morphology that was very different from the motile single cells of R. antarcticus ANT.BR isolated from Cape Royds (Madigan et al. Reference Madigan, Jung, Woese and Achenbach2000, Baker et al. Reference Baker, Riester, Skinner, Newell, Swingley and Madigan2017). Despite the high genetic similarity, our cultured organism appears to represent a novel ecotype of R. antarcticus.
Our genomic reads from R. antarcticus JLS showed no matches to our ShortBRED antibiotic resistance markers, although the reference genome does indeed include various resistance genes including numerous resistance-nodulation-cell division (RND) and major facilitator superfamily (MFS) transporters as well as multidrug efflux components. Thus, many naturally occurring ARGs are likely to be missed by standard marker searches.
The prevalence of eukaryotic sequences in the lake water metagenomes is consistent with previous reports that protists play important roles in the Taylor Valley lake communities (Lizotte et al. Reference Lizotte, Sharp and Priscu1996, Glatz et al. Reference Glatz, Lepp, Ward and Francis2006, Bielewicz et al. Reference Bielewicz, Bell, Kong, Friedberg, Priscu and Morgan-Kiss2011, Li et al. Reference Li, Podar and Morgan-Kiss2016). Microbial eukaryotes including phototrophs and mixotrophs provide prominent functions in the lake ecology (Li & Morgan-Kiss Reference Li and Morgan-Kiss2019). In our data, the eukaryotic communities of lakes Fryxell and Bonney showed three major taxa in common: G. cryophila, M. rubrum and N. limnetica. G. cryophila is a mixotrophic cryptophyte that feeds on bacteria but also conducts photosynthesis as a secondary endosymbiont alga (van den Hoff et al. Reference Van den Hoff, Bell and Whittock2020). M. rubrum is a ciliate that consumes cryptophytes but also uses the prey chloroplasts to conduct photosynthesis (kleptoplasty; Yih et al. Reference Yih, Hyung, Hae, Myung and Young2004). N. limnetica is a heterokont alga, with red alga-derived chloroplasts, and is primarily a phototroph (Kong et al. Reference Kong, Ream, Priscu and Morgan-Kiss2012). Our Lake Bonney samples also showed sequences from Chlamydomonas, a green alga that dominates some parts of the Lake Bonney water column (Lizotte et al. Reference Lizotte, Sharp and Priscu1996, Bielewicz et al. Reference Bielewicz, Bell, Kong, Friedberg, Priscu and Morgan-Kiss2011).
We note that in the water samples, smaller aquatic phototrophs were probably missed by the 0.45 μm filter; 0.20 μm filters would have been preferable but were not available in the field at the time. Even 0.20 μm filters miss important microbial community members (Brown et al. Reference Brown, Hug, Thomas, Sharon, Castelle and Singh2015). The mat samples, however, underwent no filtration, so a broader spectrum of cell sizes was captured.
It is interesting that the Lake Fryxell water shows mainly eukaryotic phototrophs whereas the mat shows mainly cyanobacteria and proteobacterial phototrophs. The mat bacteria are likely to survive a wider range of light and temperature conditions than the eukaryotes. From the standpoint of drug resistance, cyanobacteria are more likely than eukaryotes to harbour and transfer ARGs of potential bacterial pathogens. Nevertheless, protists can regulate the bacterial ARG composition in terrestrial communities (Nguyen et al. Reference Nguyen, Chen, He and Hu2020), so this factor may be of interest when assessing lake water ARG pools.
Acknowledgements
We thank Caroline Harwood for guidance regarding culturing of the mat biofilm and Emma Stuart-Bates for the biofilm culture micrograph. We especially thank Darcy Blankenhorn for expert technical support. We also thank two anonymous reviewers for their feedback.
Financial support
Fieldwork was supported by the National Science Foundation award OPP-1637708 to RMM-K. For metagenome sequencing we thank the US Department of Energy (DOE) Joint Genome Institute, a DOE Office of Science User Facility, supported by the Office of Science of the US Department of Energy Contract No. DE-AC02-05CH11231, JGI Metagenome award 1936 to JLS. Bioinformatic analysis and biofilm culture and sequencing were supported by the National Science Foundation award MCB-1923077 to JLS.
Author contributions
ST designed the bioinformatic analysis of ARGs and drafted the manuscript. GH characterized the Rhodoferax enrichment culture and contributed to the manuscript. DB ran the bioinformatic pipelines and performed statistics. RMM-K led the field expedition and collected water samples. JLS conceived the central concept, collected the mat samples and isolated DNA, mentored students and completed the manuscript.
Availability of data and materials
Sequence read files from lake water and mat samples are deposited at NCBI under SRA accession numbers SRP104818, SRP104821, SRP098041, SRP098040, SRP098042, SRP104817, SRP098044, SRP104822, SRP098050, SRP104819, SRP104820, SRP104823. Sequence read files from cultured Rhodoferax antarcticus JLS are deposited at NCBI under SRA accession number PRJNA736311.
Supplementary material
Four supplemental tables will be found at https://doi.org/10.1017/S0954102022000360.















