Introduction
The mechanically transmitted protozoan parasite Trypanosoma evansi, a causative agent of the disease called Surra, is geographically the most widely distributed member of the genus Trypanosoma and infects the widest range of mammalian hosts, including buffaloes, camels, cattle, pigs, dogs, horses and goats (Holland et al. Reference Holland, Do, Huong, Dung, Thanh, Vercruysse and Goddeeris2003; Dargantes et al. Reference Dargantes, Mercado, Dobson and Reid2009; Desquesnes et al. Reference Desquesnes, Holzmuller, Lai, Dargantes, Lun and Jittaplapong2013; Salah et al. Reference Salah, Robertson and Mohamed2015). Trypanosoma evansi parasites are transmitted by bloodsucking flies (Tabanus and Stomoxys spp.) (Hoare, Reference Hoare1972; Sumba et al. Reference Sumba, Mihok and Oyieke1998) and also by vampire bats in South America (Hoare, Reference Hoare1965). Unlike T. congolense, a haemoparasite that is strictly restricted to the host blood vessels, the absence of T. evansi in the bloodstream can still result in pathogenic symptoms from parasites that have invaded host tissue (Holland et al. Reference Holland, Do, Huong, Dung, Thanh, Vercruysse and Goddeeris2003; Desquesnes et al. Reference Desquesnes, Holzmuller, Lai, Dargantes, Lun and Jittaplapong2013). The symptoms associated with T. evansi infections differ depending on the susceptibility of the infected host and include anaemia, fever, loss of weight and productivity as well as abortion (Vickerman et al. Reference Vickerman, Myler, Stuart and Warren1993; Desquesnes et al. Reference Desquesnes, Holzmuller, Lai, Dargantes, Lun and Jittaplapong2013). In classical trypanosome infections, most of the symptoms emerge when the host immune system targets infective parasites. Parasites that die as a result of the host immune response release biologically active products that have been associated with disease symptoms (Taylor and Authié, Reference Taylor, Authié, Maudlin, Holmes and Miles2004; Antoine-Moussiaux et al. Reference Antoine-Moussiaux, Buscher and Desmecht2009). These toxins include enzymes such as trypanosome peptidases that hydrolyse host proteins, trypanosome phospholipases that hydrolyse red blood cell membranes and trypanosome trans-sialidases that act as important factors for virulence and anaemia (Tizard et al. Reference Tizard, Nielsen, Seed and Hall1978; Antoine-Moussiaux et al. Reference Antoine-Moussiaux, Buscher and Desmecht2009; Coustou et al. Reference Coustou, Plazolles, Guegan and Baltz2012; Habila et al. Reference Habila, Inuwa, Aimola, Udeh and Haruna2012). The study of these factors, and in particular peptidases, led to the idea of developing new anti-disease vaccines that target trypanosome products that are associated with disease symptoms (Authié, Reference Authié1994; Authié et al. Reference Authié, Boulangé, Muteti, Lalmanach, Gauthier and Musoke2001; Antoine-Moussiaux et al. Reference Antoine-Moussiaux, Buscher and Desmecht2009).
Several peptidases in T. brucei, T. cruzi and Leishmania spp. that belong to the S9 prolyl oligopeptidase family of clan SC serine proteases have been identified as potential drug and vaccine targets due to their unique properties (Coetzer et al. Reference Coetzer, Goldring and Huson2008; Bastos et al. Reference Bastos, Motta, Grellier and Santana2013). Members of this family of serine peptidases in T. brucei (TbbPOP) and T. cruzi (TcrPOP) hydrolyse peptides smaller than 3 kDa at the carboxyl end of proline and alanine residues (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005, Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010; Rawlings et al. Reference Rawlings, Barrett and Finn2016). Due to their catalytic properties, trypanosomal prolyl oligopeptidases have been implicated in the hydrolysis of host peptide hormones (Bastos et al. Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010). In T. cruzi infections, studies using inhibitors specific for TcrPOP prevented the parasite from infecting mammalian host cells (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005). Unlike other prolyl oligopeptidases, recombinantly expressed TcrPOP and TbbPOP are also capable of hydrolysing large substrates such as fibronectin and native collagen (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005, Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010). During T. evansi and T. brucei infections, oligopeptidase B, a serine peptidase with a preference for hydrolysing short peptides that contain di-basic residues, is released into the host bloodstream as an active peptidase that is not inhibited by host serpins where it has been implicated in hydrolysing host peptide hormones (Morty et al. Reference Morty, Lonsdale-Eccles, Mentele, Auerswald and Coetzer2001; Reference Morty, Pelle, Vadasz, Uzcanga, Seeger and Bubis2005; Coetzer et al. Reference Coetzer, Goldring and Huson2008). The abnormal degradation of host atrial natriuretic factor by oligopeptidase B during T. evansi infection in rats is associated with increased blood volume, a situation that results in various disease lesions such as cardiomyopathy and heart failure (Morty et al. Reference Morty, Pelle, Vadasz, Uzcanga, Seeger and Bubis2005). Oligopeptidase B in T. cruzi is crucial during the invasion of non-phagocytic host cells by stimulating the recruitment and fusion of lysosomes at the site of entry (Burleigh et al. Reference Burleigh, Caler, Webster and Andrews1997; Caler et al. Reference Caler, Vaena de Avalos, Haynes, Andrews and Burleigh1998). In Leishmania spp., the deletion of oligopeptidase B leads to an accumulation of membrane associated enolase, an enzyme that has been implicated as a virulence factor (Swenerton et al. Reference Swenerton, Zhang, Sajid, Medzihradszky, Craik, Kelly and McKerrow2011). The activity of TbbPOP and other closely related TbbPOP-like peptidases has been shown to be dependent on the presence of TbbOPB, where deletion of the gene for the latter leads to an increase in activity of POP-like peptidases (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012). The interdependent role that both peptidases play in host peptide hormone hydrolysis is implied when looking at the amino acid residues present in mammalian peptide hormones, with many containing prolyl and alanyl, or arginyl and leucyl moieties (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012).
To fully understand the role that serine oligopeptidases play in parasite physiology and host pathogenesis, prolyl oligopeptidase, prolyl oligopeptidase-like and oligopeptidase B null mutants (Δpop, Δpop-like and Δopb, respectively) were generated in T. evansi RoTat 1.2 parasites. Our results suggest that prolyl oligopeptidase-like serine peptidase is likely to influence host immune responses during infection by affecting interleukin concentrations in the host. These results would require further experiments using the bovine host in order to elucidate the mechanism of immune escape by T. evansi during infection.
Methods
Trypanosome culture
Bloodstream forms of T. evansi RoTat 1.2 wild-type were obtained from the Institute of Tropical Medicine, Antwerp, Belgium and were isolated in 1982 from a buffalo in Indonesia (ITMAS #020298) (Claes et al. Reference Claes, Verloo, De Waal, Urakawa, Majiwa, Goddeeris and Buscher2002). Both bloodstream form wild-type parasites and null mutant clones generated were cultured in supplemented Iscove's Modified Dulbecco's Medium (IMDM) as previously described (Hirumi and Hirumi, Reference Hirumi and Hirumi1989). Briefly, 1 L of IMDM containing 3·6 mm NaHCO3 (Thermo Fischer Scientific, Roskilde, Denmark) was supplemented with 1 mm hypoxanthine, 1 mm sodium pyruvate, 0·16 mm thymidine, 0·05 mm bathocuprone sulphate, 1·0 mm l-cysteine and 0·2 mm 2-mercaptoethanol. The pH was adjusted to between 7·2 and 7·4 and 10% (v/v) heat-inactivated fetal calf serum (Gibco, Paisley, UK) was added before filtration using a 0·2 µm filter.
Generation of serine peptidase null mutants
Cloning
Trypanosoma evansi RoTat 1.2 strain genomic DNA was extracted from in vitro cultures as previously described (Medina-Acosta and Cross, Reference Medina-Acosta and Cross1993) and used as a template for PCR. The sequences flanking the POP, POP-like and OPB genes in T. evansi were identified in the Tritryp database (http://tritrypdb.org/tritrypdb/TevansiSTIB805) and primers designed as shown in Supplementary Table S1 with the respective expected sizes of amplification products indicated and restriction sites underlined.
The six generated PCR products were cloned into the TOPO® TA cloning vector (Thermo Fischer Scientific, Roskilde, Denmark) and serially sub cloned into two knock-out vectors bearing resistance for neomycin (pGLneo) (ten Asbroek et al. Reference ten Asbroek, Ouellette and Borst1990) and blasticidin (pGLbla) (Kimura et al. Reference Kimura, Takatsuki and Yamaguchi1994) to give pGLneoTePOP, pGLneoTePOP-like pGLneoTeOPB, pGLblaTePOP, pGLblaTePOP-like and pGLblaTeOPB, where the antibiotic resistance gene was flanked by the 5′ and 3′ regions of all three genes in both knock-out vectors. Orientation of the cloned 5′ and 3′ inserts was confirmed by PCR using vector and insert specific primers.
Parameters for transfection using the 4D-Nucleofector™ system
In order to transfect T. evansi RoTat 1.2 parasites, it was necessary to define the conditions required for the successful integration of recombinant plasmids in trypanosomes when using the 4D-Nucleofector™ system (Lonza, Levallois-Perret, France). Following recommendations from the manufacturer, an optimization experiment that combines 23 different Nucleofector™ programs with 5 different solutions (P1–P5) for a total of 115 reactions was set up.
Briefly, for each reaction, a pellet of 106 parasites was resuspended in 20 µL of solutions P1–P5 along with 1 µg of NotI-linearized pLEW20-green fluorescent protein (GFP), a plasmid that was previously used for GFP expression in T. brucei (Wirtz et al. Reference Wirtz, Leal, Ochatt and Cross1999; Coustou et al. Reference Coustou, Guegan, Plazolles and Baltz2010), and transfected using one of the programs recommended by the manufacturer. Transfected parasites were first cultured for 24 h in the supplemented IMDM medium (Hirumi and Hirumi, Reference Hirumi and Hirumi1989) before addition of 2 µg mL−1 of phleomycin for the selection of viable clones. Successful clones were analysed for GFP expression using flow cytometry. The successful combination of transfection program and P1–P5 solutions from the Nucleofector™ kit was established for gene knock-out experiments.
Transfection using serine peptidase knock-out plasmids
Briefly, a pellet of 107 parasites was resuspended in 100 µL of P solution and mixed with 10 µg of NotI-linearized pGLneo plasmid (pGLneoTePOP, pGLneoTePOP-like or pGLneoTeOPB) before transfection with the program setting identified as described above. Stably transfected trypanosomes were first cultured for 24 h before addition of neomycin (2·5 µg mL−1). Single knock-out clones, generated by limiting dilution, were expanded and taken through a second round of transfection with the corresponding NotI-linearized recombinant pGLbla plasmid (pGLblaTevPOP, pGLblaTevPOP-like and pGLblaTevOPB) and selected using both neomycin (2·5 µg mL−1) and blasticidin (5 µg mL−1) as described (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012).
Confirmation of clones using Southern blot with digoxigenin-labelled probes and measurements of RNA abundance using the Affymetrix® whole transcript array
To confirm the stable transfection of both plasmids and the deletion of all three serine peptidase genes targeted in T. evansi, a Southern blot was carried out using digoxigenin probes according to the manufacturer's protocol (Roche, Mannheim, Germany). Briefly, two probes that spanned either the gene coding region (probe 1) or the external region of recombination (probe 2) were prepared by PCR incorporating digoxigenin-labelled nucleotides. Enzyme-restricted genomic DNA isolated from the selected clones (PstI for TevPOP, SmaI for TevPOP-like and DraI for TevOPB) was separated on a 1% (w/v) agarose gel (110 V for 4 h), transferred onto a DNA-binding nylon membrane (GE healthcare, Buckinghamshire, UK) using capillary action and cross-linked with UV before pre-hybridization at 42 °C and subsequent hybridization with each of the digoxigenin-labelled probes. Blots were washed with stringency buffer [0·3 m NaCl, 30 mm sodium citrate, pH 7·0 containing 0·1% (v/v) sodium dodecyl sulphate (SDS)] probed with anti-digoxigenin alkaline phosphatase (1:10 000) in Tris-buffered saline (0·1 m Tris–HCl, 0·1 m NaCl, pH 9·5) and revealed using chemiluminescent alkaline phosphatase substrate (Roche, Mannheim, Germany).
In order to further characterize the gene-deleted clones developed, RNA extracted from six replicates each of all three serine peptidase knock-out and wild-type clones was used for hybridization onto a whole transcript array for Trypanosome spp. developed by Affymetrix (Santa Clara, California, USA). RNA extraction was carried out using an extraction kit based on acid guanidinium thiocyanate (Chomczynski and Sacchi, Reference Chomczynski and Sacchi1987). Briefly, extracted RNA was processed through several cycles of amplification which include the first-strand cDNA synthesis, second-strand cDNA synthesis, in vitro cRNA synthesis and final second-cycle single-strand cDNA synthesis; all according to the manufacturer's protocol. Single-strand cDNA generated was fragmented, labelled and hybridized to the Trypanosome spp. whole transcript array before processing using the Gene Titan® Multi-Channel (MC) Instrument (Santa Clara, California, USA). Results generated where analysed using Affymetrix® Expression console software and interpreted with Affymetrix® Transcriptome Analysis Console (TAC) Software. Lists of genes were prepared using the T. brucei annotation with figures on fold change, analysis of variance (ANOVA) P-value and FDR (false discovery rate) adjusted P-value assigned to each gene described.
Enzymatic activity analysis of T. evansi serine peptidase knock-out clones
Parasites (1×107), were removed from culture, washed twice in phosphate-buffered saline (PBS), pH 7·2 and resuspended in 900 µL of 0·1% (w/v) Brij-35 in distilled water containing 10 µg mL−1 soyabean trypsin inhibitor (SBTI) to inhibit other parasite serine proteases, 10 µ m L-trans-epoxysuccinyl-L-leucylamido(4-guanidino)butane (E-64) and 1 mm ethylenediaminetetraacetic acid (EDTA). The lysate was incubated for 10 min on ice, 100 µL of 10 × PBS added and centrifuged (10 000 g , 5 min, 4 °C). The protein concentration of the lysate supernatant was determined using the bicinchoninic acid (BCA™) protein assay (Thermo Fischer Scientific, Massachusetts, USA). The hydrolysis of carboxybenzyl (Z)–Arg–Arg–7-amino-4-methylcoumarin (AMC), Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC (Bachem, Torrance, USA) by the total parasite protein extract (5 µg) was determined for each clone. Briefly, parasite protein extract diluted in 0·1% (w/v) Brij-35 was incubated with assay buffer [200 mm Tris–HCl buffer, pH 8, 10 mm dithiothreitol (DTT) and 0·02% (w/v) NaN3] for 10 min at 37 °C. Samples of the diluted parasite protein extract were combined with the appropriate fluorescent substrate (20 µ m) and the fluorescence read (excitation at 360 nm and emission at 460 nm) using a Synergy H1 microplate reader (BioTek, Vermont, USA). Lysis buffer [0·1% (w/v) Brij-35, containing 10 µ m E-64, 10 µg mL−1 SBTI, 1 mm EDTA and 1 × PBS] was used as a negative control. Parallel activity assays with inhibitors were carried out using 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) for TevOPB and 100 µ m tosyl phenylalanyl chloromethyl ketone (TPCK) for TevPOP. When determining cysteine peptidase levels in total parasite extract, hydrolysis of Z–Phe–Arg–AMC was measured using the same protocol but with 10 µ m E-64 replaced by 1 mm AEBSF and 1 µg ml−1 of pepstatin A. A parallel activity assay using 10 µ m E-64 as an inhibitor for cysteine peptidases was performed. For statistical analyses, values were expressed as means ± standard error of the mean (s.e.m.). Significance tests were calculated by using two-way ANOVA and were considered significant at a P value < 0·05.
Mouse infections
In order to observe what role the three serine peptidases, TevPOP, TevPOP-like and TevOPB play in T. evansi virulence and infection, one group of eight female, 8-week-old BALB/c mice for each gene deletion (three experimental groups housed together for each group) was inoculated by intraperitoneal injection using an insulin syringe with 1 × 104 Δpop, Δpop-like or Δopb T. evansi parasites per mouse resuspended in 50 µL of PBS. A control group was also infected with 1 × 104 T. evansi RoTat 1·2 wild-type parasites resuspended in 50 µL of PBS. Parasitaemia was measured on alternative days by bleeding from the tail and survival of mice was monitored during infection. Parasitaemia was estimated in each infected mouse using the rapid matching method as previously described (Herbert and Lumsden, Reference Herbert and Lumsden1976). Blood samples measured for parasitaemia were blinded to the readers. Plasma samples were also collected from the different groups of mice using heparinized capillary tubes over the course of infection and stored at −80 °C for further analysis. Mice were locally sourced and housed at the University of Veterinary Medicine in Vienna. Infection and care of infected mice was carried out using protocols approved by the institutional ethics committee of the University of Veterinary Medicine, Vienna and the national authority according to Section 26 of the Austrian Law for Animal Experiments, Tierversuchsgesetz 2012-TVG 2012 under the No. GZ 68·205/0069-WF/II/3b/2014.
Mouse Bio-Plex cytokine assay
A custom 10-plex Bio-Plex assay (Bio-Rad, Hercules, USA) was used to quantify the plasma levels of 10 interleukins in mouse (Mo) plasma [interferon-gamma (IFN-γ), tumour necrosis factor-alpha (TNF-α), IL-1α, IL-1β, IL-4, IL-6, IL-10, IL-12 (p40), IL12 (p70) and IL-13] as described by the manufacturer. Briefly, a 1 in 4 dilution of plasma collected at different time points from the different groups of mice was incubated with beads coupled to monoclonal antibodies specific for each component of the interleukin panel. Samples were washed before adding detection antibodies and developed for reading using the Bio-Plex® 200 suspension array system. Absolute interleukin concentrations were calculated using Bio-Plex Manager™ software.
Mouse spleen cell assay and intracellular IL-10 staining
Mouse spleen cells (106 per experiment) isolated from individual mice (3H-Biomedical, Uppsala, Sweden) were incubated with 5 × 105 parasites from TevPOP-like and T. evansi RoTat 1.2 wild-type clones in duplicate. Paired experiments were carried out for each vial of spleen cells to differentiate between different sources of mouse cells. Spleen cells were harvested 24 h after incubation and prepared for flow cytometry as previously described (Wijewardana et al. Reference Wijewardana, Kristoff, Xu, Ma, Haret-Richter, Stock, Policicchio, Mobley, Nusbaum, Aamer, Trichel, Ribeiro, Apetrei and Pandrea2013). Briefly, cells were washed in PBS (pH 7·2) and resuspended in cell surface staining mix [fluorescence-activated cell sorting (FACS) buffer: PBS pH 7·2, 2% (v/v) foetal calf serum, anti-CD3 antibody-clone 145-2C11 and anti-CD8 antibody-clone 53-6·7; BD Biosciences, New Jersey, USA] and incubated for 30 min at 4 °C. Surface stained cells were washed in FACS buffer and stained using a live/dead staining mix containing an amine reactive dye (BD Biosciences) in FACS buffer for 15 min at 4 °C. The cells were then fixed and permeabilized using BD Cytofix/Cytoperm™ (BD Biosciences) according to the manufacturer's instructions. The fixed cells were resuspended in intracellular master mix (Permeabilization buffer, IL-10 antibody-clone JES5-16E3; BioLegend, San Diego, USA) and incubated for 30 min at 4 °C. The cells were given a final wash using permeabilization buffer and resuspended in FACS buffer. The stained cells were analysed using the Gallios™ flow cytometer (Beckman Coulter, California, USA) and results evaluated using Kaluza™ software (Beckman Coulter). The Wilcoxon matched-pairs rank test was used to calculate significance values.
Statistical analysis
In order to assess the effects of parasite gene deletion on host interleukins, a regression model was developed with plasma interleukin concentration as the quantitative response variable, parasitaemia and time after infection as quantitative predictors, and the gene deleted as a qualitative predictor variable. Two-way interactions between all predictor variables were included in the model. Because interleukin measurements were made from pooled plasma in each group of mice infected, two analytical replicates of the bulked sample of all mice within one treatment at each time point were obtained and the statistical analysis was then performed on these bulked values. Since parasitaemia values were obtained for each individual mouse, the parasitaemia values were averaged for mice within one treatment per time point. The detection limit for parasitaemia was 5 × 105 mL−1 when using the rapid matching method (Herbert and Lumsden, Reference Herbert and Lumsden1976). Observations below this limit do not meet the missing-at-random assumption, and therefore leaving them out can introduce bias into the analysis. To avoid this bias, a plug-in value of half of the detection limit was used wherever parasitaemia was not detectable (Barr et al. Reference Barr, Landsittel, Nishioka, Thomas, Curwin, Raymer, Donnelly, McCauley and Ryan2006). Parasitaemia was measured daily on the same mice for the duration of the study, and therefore bulked samples in time cannot be considered independent. This serial correlation was taken into account by fitting a linear mixed model with an additional random bulk-by-time effect, with the bulked samples as subject and a serial correlation structure in time for the random effect. Analytical replicates are independent between time points and therefore no serial correlation was modelled to the residuals. The structure of the variance–covariance matrix for the bulk-by-time effect was selected based on the Akaike information criterion (AIC) (Akaike, Reference Akaike1974). To this end, fixed effects and covariance structure were simultaneously estimated with the Restricted Maximum Likelihood algorithm (Patterson and Thompson, Reference Patterson and Thompson1971). Tested were a first-order autoregressive and a compound symmetry structure. As the number of mice decreased over time, the variance of the random bulk-by-time effect was assumed to be inversely proportional to the number of mice samples in the bulk. This weighting allowed accounting for an increasing variance with a decreasing number of mice in a treatment over time. To implement the weighting using our mixed model software, we crossed the bulk-by-time effect with a continuous covariate that was equal to the inverse of the square root of the number of mice samples present in the bulk at the given time. This ensured that the variance of the random effect is inversely proportional to the number of mice samples in the bulk. Details on the weighting can be found in the Supplementary Section S2. Diagnostic plots were used to check the assumptions of normality and homoscedasticity. For the interleukin Mo IL-10 (mouse IL-10), a square root transformation was necessary to stabilize the variance. Input variable selection was then done with backward selection based on AIC, and for this purpose, models were fitted using Maximum Likelihood estimation. All analyses were performed with the MIXED procedure in SAS 9.4, and the serial correlation was accounted for using the RANDOM statement of the same procedure.
Results
Generation of T. evansi RoTat 1.2 serine peptidase null mutants
The conditions necessary for successful transfection of T. evansi RoTat 1.2 parasites using the 4D-Nucleofector™ system were established using the pLew20b-GFP plasmid. The matrix of 115 conditions recommended by the manufacturer yielded two combinations that produced viable clones: the program EF100 combined with solution P3 and the program DI100 with solution P5. Following the transfection, the potential recombinants were analysed for GFP fluorescence using flow cytometry (Supplementary Fig. S1). The combination of Program DI100 with solution P5 produced a higher number of clones when compared with program EF100 combined with solution P3 (results not shown) and was therefore chosen for future experiments.
The transfection of wild-type bloodstream forms of T. evansi RoTat 1.2 with pGLneo plasmids (pGLneoTePOP, pGLneoTePOP-like or pGLneoTeOPB) produced three distinct heterozygote clones resistant to neomycin; Δpop::NEO/POP, Δpop-like::NEO/POP-LIKE and Δopb::NEO/OPB. A second round of transfection with pGLbla plasmids (pGLblaTePOP, pGLblaTePOP-like or pGLblaTeOPB), using the respective single knock-out clones generated in the first round, resulted in the selection of three null mutant clones: T. evansi RoTat 1.2 Δpop, T. evansi RoTat 1.2 Δpop-like and T. evansi RoTat 1.2 Δopb, all of which were resistant to both blasticidin and neomycin. The deletion of TevPOP, TevPOP-like and TevOPB were confirmed by Southern blot using two probes for each deletion. Probe 1 was gene specific as shown for using TevPOP as an example, and probe 2 was an external probe that targeted a region outside the recombination region (Fig. 1A).
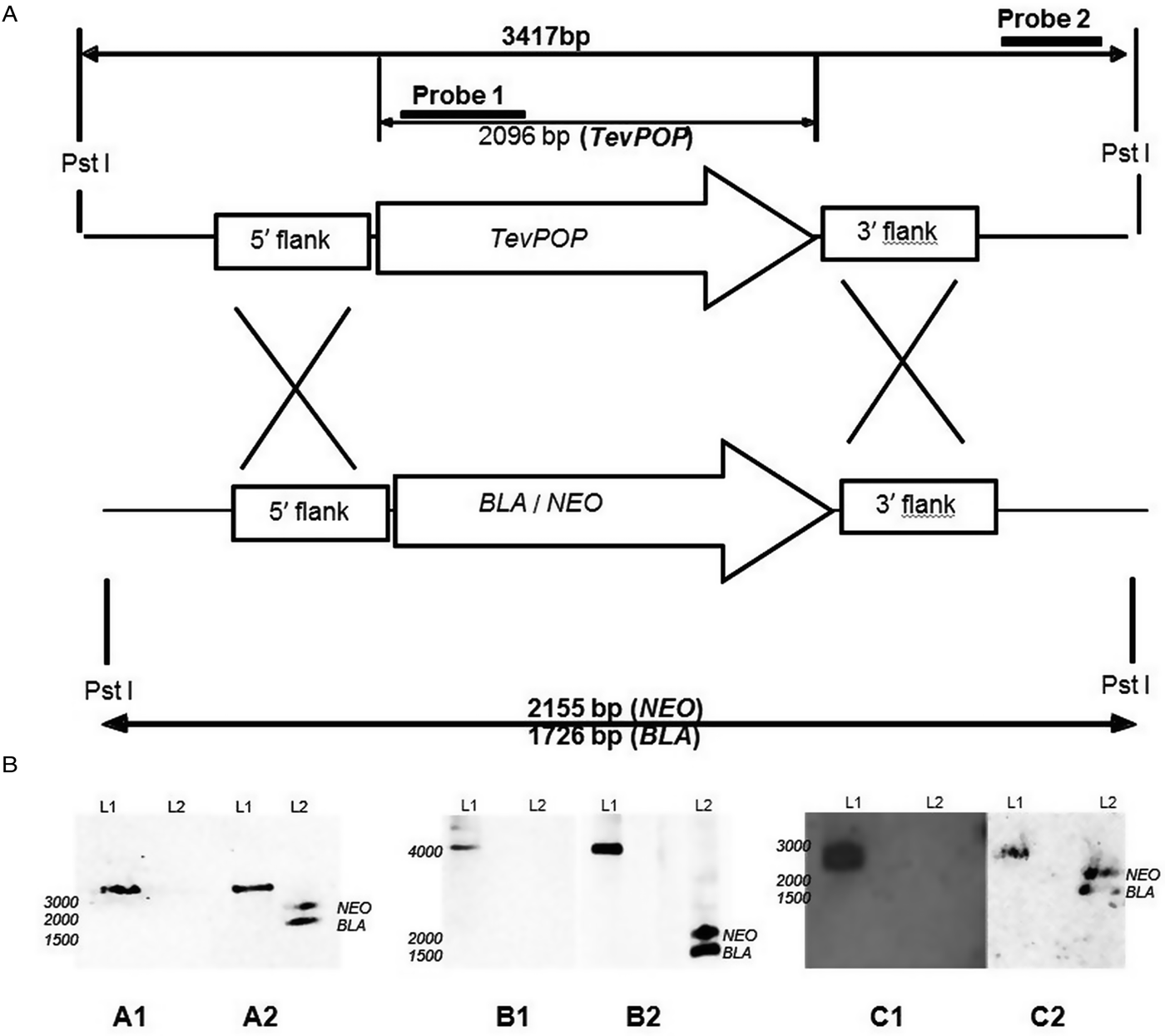
Fig. 1. Deletion of the POP, POP-like and OPB genes in T. evansi strain RoTat 1.2. (A) Schematic diagram showing the TevPOP locus. TevPOP, blasticidin (BLA) and neomycin (NEO) genes are shown by ![]() , 5′ and 3′ flanking regions by
, 5′ and 3′ flanking regions by ![]() . Probes prepared and the sizes expected on a Southern blot after restriction with PstI are in bold. TevPOP is used as a template with SmaI restriction for TevPOP-like and DraI restriction for TevOPB. (B) Southern blot of T. evansi RoTat 1.2 wild-type TevΔpop, TevΔpop-like and TevΔopb null mutants, NEO (neomycin);BLA (blasticidin). Numbers to the left indicate running positions of molecular mass markers. (A1) T. evansi RoTat 1.2 wild-type (L1) and Δpop null mutant (L2) genomic DNA incubated with probe 1 at slightly >3000 bp for TevPOP; (A2) incubated with probe 2 at slightly >3000 bp for TevPOP (L1), and at slightly >2100 bp and slightly >1700 bp for NEO and BLA respectively (L2). (B1) T. evansi RoTat 1.2 wild-type (L1) and Δpop-like null mutant (L2) genomic DNA incubated with probe 1 at slightly >4100 bp for TevPOP-like; (B2) incubated with probe 2 at >4100 bp for TevPOP-like (L1) and at >1800 bp and slightly >1400 bp for NEO and BLA, respectively (L2). (C1) T. evansi RoTat 1.2 wild-type (L1) and Δopb null mutant (L2) genomic DNA incubated with probe 1 at slightly >2800 bp for TevOPB; (C2) incubated with probe 2 at slightly >2800 bp for TevOPB (L1) and at slightly >2100 bp and slightly >1600 bp for NEO and BLA, respectively (L2).
. Probes prepared and the sizes expected on a Southern blot after restriction with PstI are in bold. TevPOP is used as a template with SmaI restriction for TevPOP-like and DraI restriction for TevOPB. (B) Southern blot of T. evansi RoTat 1.2 wild-type TevΔpop, TevΔpop-like and TevΔopb null mutants, NEO (neomycin);BLA (blasticidin). Numbers to the left indicate running positions of molecular mass markers. (A1) T. evansi RoTat 1.2 wild-type (L1) and Δpop null mutant (L2) genomic DNA incubated with probe 1 at slightly >3000 bp for TevPOP; (A2) incubated with probe 2 at slightly >3000 bp for TevPOP (L1), and at slightly >2100 bp and slightly >1700 bp for NEO and BLA respectively (L2). (B1) T. evansi RoTat 1.2 wild-type (L1) and Δpop-like null mutant (L2) genomic DNA incubated with probe 1 at slightly >4100 bp for TevPOP-like; (B2) incubated with probe 2 at >4100 bp for TevPOP-like (L1) and at >1800 bp and slightly >1400 bp for NEO and BLA, respectively (L2). (C1) T. evansi RoTat 1.2 wild-type (L1) and Δopb null mutant (L2) genomic DNA incubated with probe 1 at slightly >2800 bp for TevOPB; (C2) incubated with probe 2 at slightly >2800 bp for TevOPB (L1) and at slightly >2100 bp and slightly >1600 bp for NEO and BLA, respectively (L2).
A Southern blot using the internal gene-specific probe 1 for TevPOP revealed a slightly >3000 bp fragment (see Fig. 1A for the expected sizes of the fragments) in PstI-restricted wild-type T. evansi RoTat 1.2 genomic DNA containing full length TevPOP (Fig. 1B, A1, L1). No band was detected when the same probe was used for PstI-restricted TevΔpop null mutant genomic DNA (Fig. 1B, A1, L2). The external probe 2 also revealed a slightly >3000 bp fragment when incubated with PstI restricted wild-type RoTat 1.2 genomic DNA (Fig. 1B, A2, L1). To ensure that both resistance genes were in the right locus, a Southern blot using the external probe 2 on PstI-restricted TevΔpop null mutant genomic DNA revealed a slightly >2000 bp fragment for NEO (Fig. 2B, A2, L2) and a slightly >1600 bp fragment for BLA (Fig. 1B, A2, L2). Southern blots were also carried out for TevPOP-like and TevOPB null mutants using their respective probes. Probe 1 for TevPOP-like revealed a slightly >4000 bp band when using T. evansi RoTat 1.2 genomic DNA (Fig. 1B, B1, L1) with no band seen for Δpop-like (Fig. 1B, B1, L2). Probe 2 for TePOP-like also revealed a slightly >4000 bp band when using T. evansi RoTat 1.2 genomic DNA as expected (Fig. 1B, B2, L1), at slightly >1800 bp for NEO and slightly >1400 bp for BLA when using Δpop-like DNA (Fig. 1B, B2, L2). Probe 1 for TevOPB revealed a slightly >2800 bp band when using T. evansi RoTat 1.2 genomic DNA (Fig. 1B, C1, L1) with no band seen for Δopb (Fig. 1B, C1, L2). Probe 2 for TevOPB also revealed a slightly >2800 bp band when using T. evansi RoTat 1.2 genomic DNA (Fig. 1B, C2, L1), and at slightly >2100 bp for NEO and 1675 bp for BLA when using Δopb DNA (Fig. 1B, C2, L2).
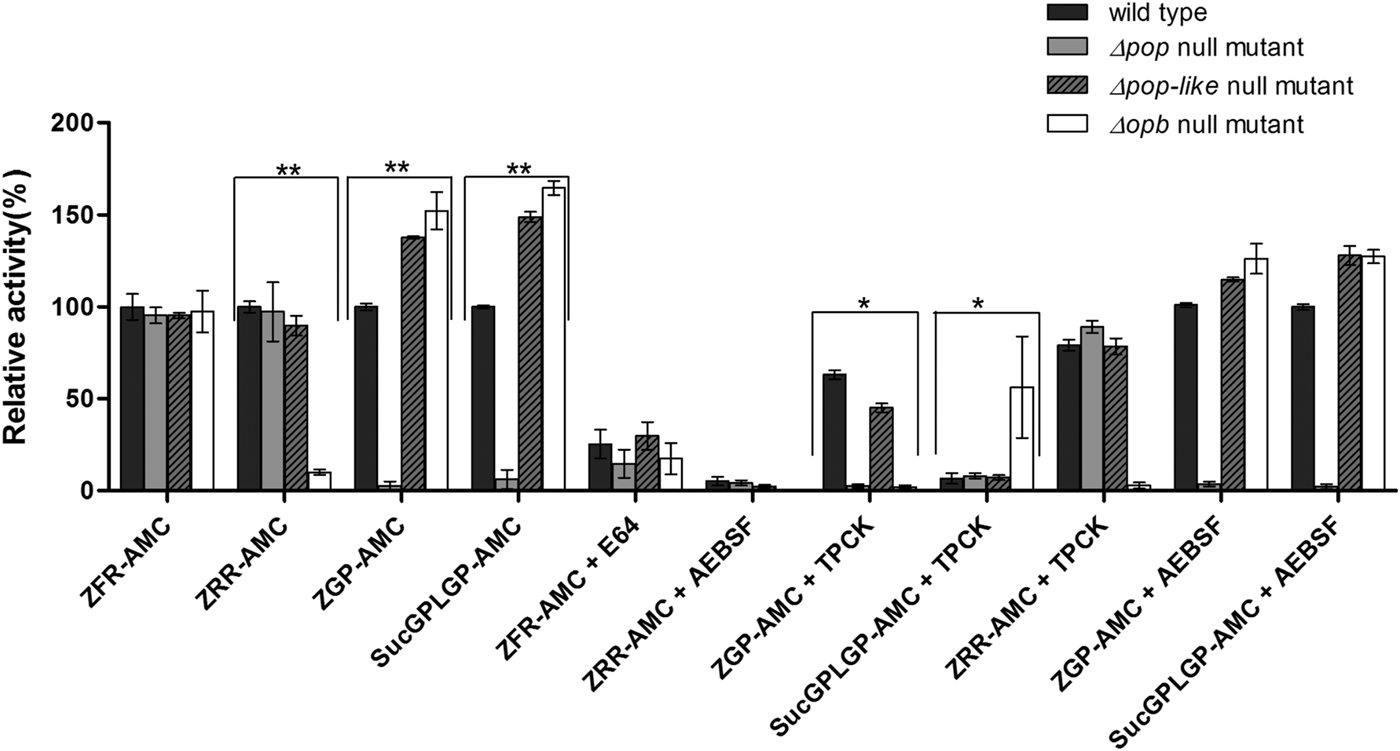
Fig. 2. Enzymatic characterization of null mutant parasites. The hydrolysis of OPB substrates, Z–Arg–Arg–AMC (ZRR–AMC) and Z–Phe–Arg–AMC (ZFR–AMC) and POP substrates Z–Gly–Pro–AMC (ZGP–AMC) and Suc–Gly–Pro–Leu–Gly–Pro–AMC (SucGPLGP–AMC) by lysates of Δopb null mutant, Δpop null mutant and Δpop-like null mutant and T. evansi RoTat 1.2 wild-type parasites was assessed with or without specific inhibitors E64, AEBSF or TPCK included in the assay. Data are presented as means ± s.e.m. (n = 3) of fluorescence values plotted relative to the wild-type as a percentage for each experiment, with no lysate fluorescence deducted from each value as background. **P < 0·001 and *P < 0·05 were calculated in comparison to the wild-type.
Gene knock-out clones along with wild-type controls were also analysed using the Affymetrix whole transcript array specific for Trypanosome spp. Each group of gene deletion replicate clones were analysed using the T. evansi RoTat 1.2 wild-type clone as a positive control and gene lists generated using T. brucei TREU927 annotation. The top threefold change targets detected were Tb927·11·12850 (oligopeptidase B), Tb927·5·4300 (prolyl oligopeptidase-like) and Tb927·10·8020 (prolyl endopeptidase) with fold changes of −146·61, −108·62 and −104·02, respectively (Supplementary Table S2). This confirms that the targeted genes were correctly deleted in the experiment. The downregulation or upregulation of other genes observed in the array such as Tb927·2·170 (leucine-rich repeat protein) and Tb927·10·10240 (procyclin-associated gene 1 protein) are associated with variant surface glycoprotein expression, which switches periodically during an infection and as expected show a different expression profile when compared with the parasite clone used for annotation.
The growth of parasites in vitro was also analysed with no significant differences in replication observed between the different gene knock-out clones when compared with the wild-type (Supplementary Fig. S3).
Activity assays using serine peptidase null mutant clones
Lysates of T. evansi RoTat 1.2 Δpop, Δpop-like and Δopb null mutants and wild-type parasites were tested for serine peptidase activity using four substrates; Z–Phe–Arg–AMC, Z–Arg–Arg–AMC, Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC. Whereas Z–Arg–Arg–AMC is the preferred substrate for oligopeptidase B hydrolysis (Coetzer et al. Reference Coetzer, Goldring and Huson2008), Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC are both used for measuring prolyl oligopeptidase activity with Suc–Gly–Pro–Leu–Gly–Pro–AMC appearing to be a better substrate than Z–Gly–Pro–AMC (Bastos et al. Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010).
All the mutant and wild-type parasites were able to hydrolyse Z–Phe–Arg–AMC, a substrate routinely used for measuring the activity of parasite cysteine peptidases and used as a control in this experiment (Mbawa et al. Reference Mbawa, Gumm, Shaw and Lonsdale-Eccles1992; Caffrey et al. Reference Caffrey, Hansell, Lucas, Brinen, Alvarez Hernandez, Cheng, Gwaltney, Roush, Stierhof, Bogyo, Steverding and McKerrow2001). Wild-type T. evansi RoTat 1.2 parasites readily hydrolysed Z–Arg–Arg–AMC (expressed as 100% activity), while the Δpop and Δpop-like null mutants displayed similar activities at equal and approximately 90% of that of the wild-type, respectively (Fig. 2). The Δopb null mutant parasites were unable to hydrolyse Z–Arg–Arg–AMC and gave comparable values to the no-lysate control as expected (Fig. 2). The substrates Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC were used to characterize the Δpop null mutant clones and as expected were unable to hydrolyse either substrates (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005, Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010). Interestingly, Δpop-like mutant clones retained their ability to hydrolyse both Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC substrates with a significantly higher activity (Fig. 2). A similar trend is seen with the T. evansi Δopb null mutant, a phenomenon that has previously been observed in oligopeptidase B null mutants when using Z–Gly–Pro–AMC as a substrate (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012)
The effect of class specific inhibitors was also included in the activity assay in order to further characterize the activity of the mutant clones generated. The inhibitors E-64 and AEBSF effectively inhibited the hydrolysis of Z–Phe–Arg–AMC and Z–Arg–Arg–AMC respectively in all mutant and wild-type parasites (Fig. 2). The effect of TPCK on the hydrolysis of Z–Gly–Pro–AMC by wild-type and Δpop-like mutant parasites was incomplete when compared with Δopb null mutants (Fig. 2). This effect is however reversed when using Suc–Gly–Pro–Leu–Gly–Pro–AMC as a substrate where Δopb null mutants retain activity in the presence of TPCK when compared with wild-type and Δpop-like mutant parasites (Fig. 2). When Z–Arg–Arg–AMC hydrolysis was performed with TPCK and Z–Gly–Pro–AMC or Suc–Gly–Pro–Leu–Gly–Pro–AMC with AEBSF, all mutant and wild-type parasites displayed profiles similar to when hydrolysis was carried out without inhibitors (Fig. 2).
Evaluation of the T. evansi RoTat 1.2 Δpop, Δpop-like and Δopb null mutants in mice
Mice were inoculated with the three different T. evansi mutants in parallel with wild-type parasites in order to observe their in vivo dynamics. T. evansi RoTat 1.2 wild-type infections in mice were significantly virulent with a pre-patent period of 2 days (Fig. 3A) and a median survival rate of 11 days (Fig. 3B). The group of mice infected with T. evansi Δpop-like null mutant parasites survived longer than mice infected with control T. evansi RoTat 1.2 wild-type parasites with a median survival rate of 15 days compared with 11 days in the control group (Fig. 3B). Mice infected with T. evansi Δpop-like null mutant parasites were also able to survive the first wave of parasitaemia when compared with the wild-type control, although the parasitaemia profiles in terms of numbers were similar to the wild-type control (Fig. 3A). The two groups of mice infected with either T. evansi Δpop or T. evansi Δopb null mutant parasites did not survive significantly longer than the control group with median survival rates of 9 and 7 days, respectively (Fig. 3B), and did not recover from the first wave of parasitaemia (Fig. 3A).

Fig. 3. Mice infected using T. evansi Δpop-like null mutants survive significantly longer when compared with groups infected with the wild-type parasites or other mutants generated in this study. (A) Parasitaemia in mice infected with T. evansi RoTat 1.2 wild-type parasites compared with Δopb null mutant-, Δpop null mutant- and Δpop-like null mutant infected mice. Each value is a mean of 8 mice. Intraperitoneal infections were carried out using 1 × 104 parasites per mouse. (B) Kaplan–Meier survival analysis for mice infected with T. evansi RoTat 1.2 wild-type parasites compared with Δopb null mutant, Δpop null mutant and Δpop-like null mutant (n = 8 in each group) *P < 0·05 in comparison to the wild-type.
Comparison of interleukin levels in mice inoculated with T. evansi RoTat 1.2 Δpop, Δpop-like, Δopb and wild-type parasites
Plasma collected from each group of infected mice over different days of infection was measured for ten different interleukin levels (Supplementary Fig. S2). For all interleukin concentrations, the effect of parasitaemia and its interaction terms was non-significant and therefore this variable was dropped from the models. Furthermore, non-significant changes in interleukin concentration over time were observed in mouse blood; with the exception of IL-10 and IL-1b (Fig. 4) where both time after infection, parasite clone and the interaction between these two variables were significant. The significant interaction term indicates that the interleukin concentration of IL-10 and IL-1b evolves differently over time for the various parasite types.

Fig. 4. Plasma analysis for IL-10 and IL-1b. (A) Plasma collected from a group of eight mice infected using T. evansi RoTat 1.2 wild-type parasites per mouse was measured for different interleukin concentrations including IL-10 and IL-1b that displayed a significant difference and plotted as pg mL−1 on the secondary Y-axis; (B) Interleukin measurements in plasma collected from mice infected with Δpop-like null mutant parasites; (C) Interleukin measurements in plasma collected from mice infected with Δpop null mutant parasites; (D) Interleukin measurements in plasma collected from mice infected with Δopb null mutant parasites. Intraperitoneal infections were carried out using 1 × 104 parasites per mouse. The lower limits of detection using luminex were 61·524 pg mL−1 for IL-10 and 112·732 pg mL−1 for IL-1b. Any measurements that fell below these limits were plotted as 0 in the graphs above.
The plasma levels of IL-10 in T. evansi RoTat 1.2 wild-type infections increased significantly over time (slope estimate of 4·8, P < 0·001, Table 1) and were highest before the mice succumbed to disease (Fig. 4A). The plasma levels of IL-1b in mice inoculated with wild-type parasites also increased significantly as the infection progressed (slope estimate of 3·401, P < 0·05, Table 2), although this was not as dramatic as the increase in IL-10 plasma concentrations.
Table 1. Parameter estimates for the fixed effects of the linear mixed model for Interleukin IL-10 concentration in mice infected with gene deleted parasites, obtained with Restricted Maximum Likelihood and a random bulk-by-time effect weighted for the decreasing number of surviving mice, and significance tests for the slope estimates for the interaction term time after infection and parasite type *P < 0·05, **P < 0·001
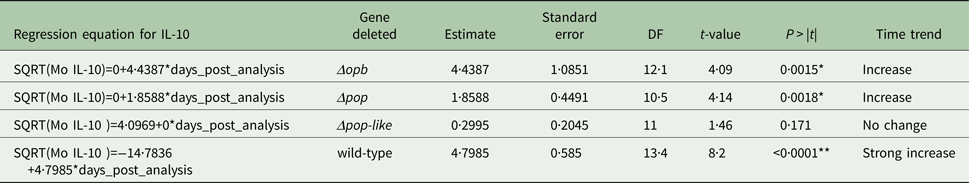
Table 2. Parameter estimates for the fixed effects of the linear mixed model for Interleukin IL-1b concentration in mice infected with gene deleted parasites, obtained with Restricted Maximum Likelihood and a random bulk-by-time effect weighted for the decreasing number of surviving mice and significance tests for the slope estimates for the interaction term time after infection and parasite type *p < 0·05
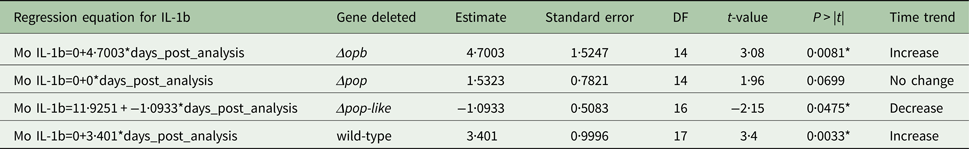
A different scenario was observed in the group of mice infected using T. evansi Δpop-like null mutants, where the concentration of IL-10 in plasma did not change significantly as infection progressed (Fig. 4B). Statistical analysis confirmed this trend with a P-value of 0·171 (Table 1). Interestingly, however, IL-1b interleukin values dropped significantly as infection with Δpop-like null mutants progressed (slope estimate −1·09, Table 2, Fig. 4B), with a P-value of 0·04 whereas wild-type infections showed a significant positive slope (estimate 3·4, P-value <0·05, Table 2).
Infections using Δpop and Δopb null mutants followed a similar trend to infections using T. evansi wild-type parasites (Figs. 4C and D respectively). In both of these infections, IL-10 production increased along with IL-1b as disease progressed (Tables 1 and 2).
In vitro analysis of IL-10 production in mouse spleen cells
In order to confirm the effect of reduced IL-10 production in mice, naïve BALB/c mouse spleen cells were incubated in paired experiments using both T evansi wild-type and TevΔpop-like parasites. The characerization of the incubated spleen cells using flow cytometry revealed that CD3+ T lymphocytes are responsible for the production of IL-10 in mouse spleens (Fig. 5A). A total of ten replicate experiments using different vials of mouse spleen cells in paired experiments consistently showed that T. evansi RoTat1.2 wild-type parasites elicited a higher percentage of T cells producing IL-10 when compared with experiments using TevΔpop-like parasites. Statistical analysis using the Wilcoxon matched-pairs signed rank test confirmed this trend giving a P value of 0·002 when ranking wild-type parasites with TevΔpop-like clones and a P value of 0·04 when compared with cells incubated without parasites (Fig. 5B).

Fig. 5. Flow cytometry analysis of naïve mouse spleen cells (A) Representative flow cytometry plots illustrating the gating strategy used to define IL-10 producing CD3+ T lymphocytes (blue) versus Non-T lymphocytes (green). (B) Wilcoxon matched-pairs signed rank test reveals a significant difference between cells incubated with wild-type parasites when compared with spleen cells incubated with Δpop-like clones at a P value of 0·002. Differences between spleen cells incubated with Δpop-like and non-incubated cells are not significant.
Discussion
The role of parasite prolyl oligopeptidases (POPs) in host physiology is closely linked to the catalysis of mammalian peptides during infection (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005; Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010; Reference Bastos, Motta, Grellier and Santana2013; Kaszuba et al. Reference Kaszuba, Rog, Danne, Canning, Fulop, Juhasz, Szeltner, St Pierre, Garcia-Horsman, Mannisto, Karttunen, Hokkanen and Bunker2012; Fajtová et al. Reference Fajtová, Štefanić, Hradilek, Dvořák, Vondrášek, Jílková, Ulrychová, McKerrow, Caffrey, Mareš and Horn2015). Although POPs from different species share similar three-dimensional structures, divergences have been observed between species in terms of inhibition and the specificity of substrates that they hydrolyse (Kaszuba et al. Reference Kaszuba, Rog, Danne, Canning, Fulop, Juhasz, Szeltner, St Pierre, Garcia-Horsman, Mannisto, Karttunen, Hokkanen and Bunker2012; Bastos et al. Reference Bastos, Motta, Grellier and Santana2013). Whereas recombinant POPs from T. brucei and T. cruzi are capable of hydrolysing proline rich native collagen, recombinant POP from Schistosoma mansoni is unable to hydrolyse substrates such as human collagens type I and IV (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005, Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010; Fajtová et al. Reference Fajtová, Štefanić, Hradilek, Dvořák, Vondrášek, Jílková, Ulrychová, McKerrow, Caffrey, Mareš and Horn2015). The exact physiological functions of POPs are yet to be fully understood, with functions seemingly related to the infective species. These divergences in function are hinted at even within the same species, with a twofold increase in the expression of procyclic POP in T. brucei compared with bloodstream forms of the parasite (Bastos et al. Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010). In this study, we observed a possible role for a POP-like peptidase from T. evansi in terms of affecting the concentrations of interleukin 10 in plasma during infection in mice. A recent study that screened 20 serine peptidase genes using RNA interference, nine of which belong to the S9 family, described an essential role for a putative type-I signal peptide peptidase (Moss et al. Reference Moss, Brown, Hamilton, Van der Veken, Augustyns and Mottram2015). Oligopeptidase B, putative POP and prolyl oligopeptidase-like from T. b. brucei (TbbOPB, TbbPOP and TbbPOP-like; Gene IDs Tb927·11·12850, Tb927·10·8020 and Tb927·5·4300, respectively) were all included in the screen. A reduction of approximately between 75% for TbbPOP and 25% for TbbPOP-like did not display any phenotype both in vitro and during mouse infections. This was, however, not unexpected, as earlier RNAi experiments targeting TbbPOP with a knockdown efficiency of up to 80% showed no effect on parasite viability (Bastos et al. Reference Bastos, Motta, Charneau, Santana, Dubost, Augustyns and Grellier2010). The deletion of TbbOPB was also previously shown to have no effect on the viability or virulence of T. brucei 427 Lister parasites (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012). The present study showed that the deletion of the TevPOP-like gene has a different effect on T. evansi parasites when used for mouse infections, with the group of mice infected able to survive the first wave of parasitaemia and with a longer median survival rate of 15 days when compared with 11 days in the wild-type infection control. Interestingly, the generation of Δopb and Δpop-like null mutants in T. evansi parasites resulted in an increase in the hydrolysis of Z–Gly–Pro–AMC and Suc–Gly–Pro–Leu–Gly–Pro–AMC, both of which are substrates used to assay for recombinant T. b. brucei POP (Bastos et al. Reference Bastos, Grellier, Martins, Cadavid-Restrepo, de Souza-Ault, Augustyns, Teixeira, Schrével, Maigret, da Silveira and Santana2005). The increase in activity for Z–Gly–Pro–AMC when the gene for oligopeptidase B is deleted has been observed in T.b. brucei Δopb null mutants (Kangethe et al. Reference Kangethe, Boulange, Coustou, Baltz and Coetzer2012). When class specific inhibitors were used to assay the activity of the mutants generated in the present study, it was shown that Δpop-like parasites retained approximately half their activity when incubated with TPCK before the incubation with Z–Gly–Pro–AMC, similar to wild-type parasites. Interestingly, this activity is inhibited in Δopb parasites, which instead shows residual activity when using Suc–Gly–Pro–Leu–Gly–Pro–AMC as a substrate after incubation with TPCK. This would imply that the increase in POP activity seen when deleting TevOPB can be attributed to TevPOP, whereas deleting TevPOP-like affects the activity of another unknown peptidase with POP activity.
Plasma collected from mice over the course of infection with all three null mutants and the wild-type control was measured for ten different interleukin concentrations (Supplementary data Fig. S2). Statistical analysis of absolute interleukin concentrations in correlation with parasitaemia revealed that IL-10 levels in mice infected using T. evansi Δpop-like mutants did not significantly change when compared with the wild-type control, whereas IL-1b levels gradually declined but with less significance when compared with wild-type controls. In order to further study the effect of deleting the TevPOP-like gene on IL-10 production, an in vitro assay using BALB/c mouse splenocytes showed that CD3+ T-lymphocytes are the major producers of IL-10 and their ability to secrete IL-10 is reduced when incubated with Δpop-like parasites as opposed to the wild-type strain. During Leishmania mexicana infections in C57BL/6 mice, T cell-derived IL-10 was observed during the chronic phase of infection as opposed to classical immune suppression by IL-10 producing macrophages or dendritic cells (Bastos et al. Reference Bastos, Motta, Grellier and Santana2013). These IL-10-producing T cells could be CD4+CD25+and FoxP3+ regulatory T cells, which suggests that wild-type parasites induce immune suppression at the onset of the infection. When Δpop-like null mutants were incubated with mouse splenic cells in vitro, a reduction in IL10 expression suggests that POP-like protease may play a role in regulating the production of the interleukin in host cells. Future experiments are required to ascertain which sub-type of T cells produce regulatory IL-10 and further elucidate which role parasite POP-like protease plays in host IL-10 production. Other parasite peptidases that have also been shown to inhibit Th1 responses during infection include a cysteine protease B from Leishmania mexicana and cathepsin B from Leishmania chagasi, that cleave human TGF-β, a cytokine that regulates IL-10 and thereby affecting immunosuppression (Somanna et al. Reference Somanna, Mundodi and Gedamu2002; Buxbaum et al. Reference Buxbaum, Denise, Coombs, Alexander, Mottram and Scott2003; Buxbaum, Reference Buxbaum2015).
In classical trypanosome infections, initial pro-inflammatory type I immune responses mediated by IFN-γ, nitric oxide (NO) and TNF are necessary for controlling the first wave of parasitaemia (Namangala et al. Reference Namangala, Noel, De Baetselier, Brys and Beschin2001b; Stijlemans et al. Reference Stijlemans, Guilliams, Raes, Beschin, Magez and De Baetselier2007; Baral, Reference Baral2010). Interleukin 1b is also a strong mediator of inflammatory immune responses during T. brucei infections (Nyakundi et al. Reference Nyakundi, Crawley and Pentreath2002). A sustained type I response is however deleterious to the mammalian host and type II non-inflammatory responses mediated by IL-4 and -10 are required later during infection to prevent immune mediated pathologies (Namangala et al. Reference Namangala, De Baetselier, Noel, Brys and Beschin2001a; Antoine-Moussiaux et al. Reference Antoine-Moussiaux, Magez and Desmecht2008; Kato et al. Reference Kato, Alibu, Nanteza, Mugasa and Matovu2015). In contrast, T. evansi infections in C57BL/6 mice do not require a type I inflammatory response in order to control parasitaemia with the role of IgM proven to be more crucial (Baral et al. Reference Baral, De Baetselier, Brombacher and Magez2007). TNF, TNF receptor gene, IFN-γ and Inducible nitric oxide synthase (iNOS) groups of knockout mice infected with T. evansi parasites all developed parasitaemia and survived at a similar rate as wild-type C57BL/6 mice suggesting that the main determinants for inflammatory response were not crucial for survival and parasitaemia in mice (Baral et al. Reference Baral, De Baetselier, Brombacher and Magez2007). When B-cell deficient and IgM deficient groups of mice were used for T. evansi infection studies, the loss of first peak parasitaemia control and subsequent rapid death was observed when compared with wild-type C57BL/6 mice. The passive transfer of IgM antibodies to B cell deficient mice restored parasite control indicating a significant contribution to protection (Baral et al. Reference Baral, De Baetselier, Brombacher and Magez2007).
The role of non-inflammatory IL-10 mediated responses during T. evansi infections has been studied in mice and cattle (Mekata et al. Reference Mekata, Konnai, Mingala, Abes, Gutierrez, Dargantes, Witola, Inoue, Onuma, Murata and Ohashi2012, Reference Mekata, Murata, Mingala, Ohashi and Konnai2015). Mice infected with T. evansi strain L2 showed a strong up-regulation of IL-10 and CCL8 which were responsible for the expansion of regulatory dendritic cells (DCs) during the acute phase of parasite infection (Mekata et al. Reference Mekata, Konnai, Mingala, Abes, Gutierrez, Dargantes, Witola, Inoue, Onuma, Murata and Ohashi2012). Inoculation of mice with regulatory DCs significantly prolonged their survival rate after infection when compared with untreated controls (Mekata et al. Reference Mekata, Konnai, Mingala, Abes, Gutierrez, Dargantes, Witola, Inoue, Onuma, Murata and Ohashi2012). This indicated that regulatory DCs inhibited the production of pro-inflammatory cytokines associated with acute pathology during T. evansi infections in mice. The upregulated expression of CCL8 and IL-10 has also been detected in cattle experimentally infected with T. evansi parasites (Mekata et al. Reference Mekata, Murata, Mingala, Ohashi and Konnai2015).
Our model for infection shows that BALB/c mice infected with either TevΔopb or TevΔpop parasites does not change the course of infection when compared with infections initiated using T. evansi RoTat 1.2 wild-type parasites. T. evansi Δpop-like mutants are also able to infect mice in a similar manner to wild-type parasites although the mice infected are able to survive longer and are associated with a lower expression of IL-10 during infection. We do however need to stress that the altered interleukin levels observed in this study could be as a result of other factors other than the deletion of POP-like peptidase.
In conclusion, we were able to generate T. evansi Δpop, Δpop-like and Δopb null mutant parasites and show that mice infected using T. evansi Δpop-like parasites were able to survive longer when compared with wild-type infections and were associated with lower IL-10 production. The association observed between Δpop-like parasites and IL-10 was also observed using a splenocyte in vitro assay. It must be emphasized that in order to fully understand the role of POP-like peptidase during T. evansi infections, experiments need to be carried out in the natural host where a more natural chronic infection is possible. This is particularly important in order to further elucidate the role of POP-like peptidase during the course of parasite infection.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/pao.2017.20
Acknowledgements
We thank Jeremy C. Mottram from the Wellcome Trust Centre for Molecular Parasitology at the University of Glasgow for providing the knock-out vectors and Philippe Büscher from the Institute of Tropical Medicine in Antwerp for providing the T. evansi Rotat 1.2 strain.
Financial Support
This work was supported by The African Renaissance Fund.
Conflicts of Interest
The authors have nothing to disclose.











