INTRODUCTION
In April 2009 a new strain of influenza A(H1N1) (‘Swine flu’) was detected in Mexico and the USA and quickly spread around the globe [1]. The new virus showed sustained human transmission with higher reproduction ratio than seasonal flu [Reference Fraser2] but the pandemic was generally characterized by mild illness and a severity of disease intermediate between seasonal flu and highly pathogenic influenza viruses, i.e. H5N1 viruses.
In the initial stages of the UK influenza A(H1N1) outbreak (April–July 2009) a containment strategy was adopted where symptomatic patients were investigated for the new virus, treated with antivirals and prophylaxis given to contacts. During this time the West Midlands experienced more cases of influenza A(H1N1) than most other UK regions [3].
By July 2009, when the UK moved from the ‘containment’ to the ‘treatment’ phase (only confirmed H1N1 patients treated), the Health Protection Agency (HPA) (Public Health) laboratory (Birmingham) had received around 11 000 specimens and confirmed over 2500 cases of influenza A(H1N1) [Reference Scriven4]. To cope with the vastly increased workload, the laboratory adopted the strategy of testing for influenza viruses only instead of testing for all other viral respiratory pathogens [Reference Scriven4].
After the initial weeks, a large proportion of the specimens received for influenza A(H1N1) testing were taken from the community rather than a hospital population. As, under normal circumstances, the laboratory receives very few specimens from a non-hospital setting this large collection of community respiratory specimens provided a unique opportunity to investigate for other respiratory viruses circulating in the community during the influenza A(H1N1) outbreak.
Many viruses other than influenza can cause respiratory tract infections with various morbidity and mortality rates worldwide. Acute viral respiratory infections are the most common infections in man, averaging two or more infections per year per person [Reference Mahony5–Reference Heikkinen and Jarvinen7]. Symptoms can often overlap with those of mild influenza and include pharyngitis, rhinorrhoea, cough, sneezing, fever, malaise and myalgia [Reference Heikkinen and Jarvinen7]. The commonest respiratory viruses other than influenza include rhinoviruses, coronaviruses, respiratory syncytial viruses (RSV), parainfluenza viruses, adenoviruses, enteroviruses, human metapneumovirus (HMPV) and human bocavirus (HboV) [Reference Heikkinen and Jarvinen7, Reference Ruohola8] most of which can cause upper and lower respiratory tract infections [Reference Mahony5].
The aims of this study were to establish which respiratory viruses were co-circulating with influenza H1N1 during the summer of 2009 and to investigate whether patients with influenza A(H1N1) had co-infections with any other respiratory viruses and compare the rate of respiratory virus co-infections in specimens from hospitalized and community patients.
METHODS
Specimen selection
Specimens (nose and/or throat swabs in transport media) received at the HPA (Public Health) laboratory (Birmingham) for influenza A(H1N1) investigation during the summer 2009 influenza A(H1N1) outbreak (sample dates between 20 June 2009 and 2 August 2009) were sorted by location and divided into two groups defined as ‘Community’ or ‘Hospital’ locations. Community specimens were from patients presenting with flu-like illness to Birmingham and District General Practitioner Emergency Rooms (BADGER) clinics. Hospital specimens were from patients admitted to hospitals in the UK West Midlands Region and having respiratory symptoms including severe flu-like illness, pneumonia, and acute respiratory failure.
Specimens were also sorted by influenza A(H1N1) PCR test results (positive or negative) into four study groups: hospital influenza A(H1N1) positive, hospital influenza A(H1N1) negative, community influenza A(H1N1) positive and community influenza A(H1N1) negative specimens.
Within each of the four study groups specimens were sorted by age of patients at sample collection date to ensure that specimens selected would represent all age groups.
Any duplicate specimens from the same patient (within 1 week of another specimen) were excluded.
Nucleic acid extraction
Specimens stored at −20°C were thawed to room temperature and vortex mixed. All samples were spiked with control brome mosaic virus (BMV) RNA to detect specimens inhibitory to PCR or extraction failure [Reference Ferns and Garson9]. All samples were tested for human RNase P to assess sample quality [Reference Dong10]. Total nucleic acid was extracted using a QIAamp One-For-All Nucleic Acid kit (Qiagen, USA) on a BioRobot MDX (Qiagen) platform running the QIAamp One-For-All MDX cV60 protocol.
Detection of viral nucleic acids
Respiratory virus detection was performed using real-time PCR. All thawed specimens were tested for targets specific for influenza A (generic for all type A influenza viruses) [Reference Carr11], influenza A (H1 specific) [12], influenza B [Reference Gunson, Collins and Carman13], RSV (generic for types A and B) [Reference Gunson, Collins and Carman13], HMPV [Reference Kuypers14], parainfluenzas 1 and 4 [personal communication, R. Gunson, West of Scotland Specialist Virology Centre, Glasgow, UK], parainfluenza 2 [Reference Cannon15], and parainfluenza 3 [Reference Gunson, Collins and Carman13], rhinovirus [Reference Gunson, Collins and Carman13], adenovirus [Reference Heim16] and coronaviruses (group 1, group 2 and SARS). Specimens with equivocal results for H1N1 by the generic influenza A- and H1-specific PCRs were tested by an N1-specific PCR [17] for confirmation of results.
Reverse transcriptase–PCR (RT–PCR) reactions for influenza, RSV, HMPV, parainfluenza and rhinovirus targets were set up in 30-μl volumes using the Invitrogen SuperScript® III Platinum® One-Step Quantitative RT-PCR system with ROX (Invitrogen, USA). Each 30 μl reaction included 10 μl nucleic acid extract template, 15 μl 2× reaction mix with ROX (containing 0·4 mm of each dNTP, 6 mm MgSO4, and 1 μm ROX) and 1 μl SuperScript III RT/Platinum Taq Mix (Invitrogen) with appropriate primers and probes and PCR grade water to 30 μl.
PCR reactions for adenovirus detection were set up in 30-μl volumes using Invitrogen TaqMan® Fast Universal PCR Master Mix (2×). Each 30-μl reaction included 3 μl nucleic acid extract template, 15 μl TaqMan® Fast Universal PCR Master Mix (2×), 0·1 μl BMV cDNA [prepared in-house and titrated to give a consistent cycle threshold (Ct) value of 30], appropriate primers and probes and PCR grade water to 30 μl.
Reactions for influenza, RSV, HMPV, parainfluenza, rhinovirus and adenovirus targets were set up in 96-well plate format and run on the Applied Biosystems 7500 Fast real-time PCR system (Applied Biosystems, USA) using the following cycling conditions: for RT-PCR; 50°C for 15 min, 95°C for 2 min followed by 45 cycles of 9°C for 15 s and 6°C for 40 s or for adenovirus PCR; 95°C for 20 s followed by 50 cycles of 95°C for 3 s, 55°C for 10 s and 65°C for 30 s.
RT-PCR reactions for coronavirus detection were set up in 25 μl volumes with 0·8 μl Invitrogen Superscript III Platinum one-step enzyme, 20 μl Master Mix, 5 μl nucleic acid extract and the following primers at 400 nm:
-
Cor F generic: ATGGGTTGGGAYTATCCXAARTGTGA,
-
Cor R1 (gp 2): GCAGTAGTTGCATCACCACTRCTAGT,
-
Cor R2 (gp 1): GCTGTACTAGCRTCACCAGAAGT,
-
Cor R3 (gp 1): GCTGTAGTTGCRTCACCAGAAGT,
-
Cor R4 (SARS): AGCAGTTGTAGCATCACCGGATGAT,
and probes at 80 nm:
-
Gp 1 probe: FAM-TTRGGYTCTAAGCATGTYA-MGBNFQ,
-
Gp 2 probe: VIC-CTTGCGAATGAATGYGC-MGBNFQ,
-
SARS probe: CY5-CAGGTTAGCTAACGAGTGTGCGCAAGTA-BHQ3.
Amplification reactions and detection of PCR products were performed using the Rotorgene™ RT-PCR system with the following cycling conditions: 50°C for 30 min, 95°C for 2 min, 45 cycles at 95°C for 15 s and 60°C for 1 min.
For each PCR reaction set up, appropriate positive control extracts for each target prepared from known positive specimens and diluted to give a consistent Ct value or, for coronavirus, also made from a pool of recombinant pUC57 plasmids containing the PCR target sequences (www.genscript.com), were included alongside negative controls from the extraction run and negative controls using PCR grade water.
RESULTS
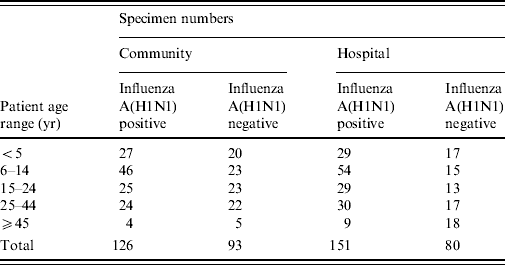
A total of 450 respiratory specimens were selected and retrieved from storage. The number of specimens by age of patient, community or hospital location and influenza A(H1N1) positivity or negativity is shown in Table 1.
Table 1. Number of specimens in each study group
No influenza A strains other than H1N1 were detected in any specimen and none of the specimens was positive for influenza B. Of the community specimens positive for influenza A(H1N1), 6·3% (8/126) had another respiratory virus detected. Of the hospital specimens positive for influenza A(H1N1), 5·3% (8/151) had another respiratory virus detected (Fig. 1). There was no statistically significant association between respiratory virus co-infection and patient location (P=0·79 Fisher's exact test).
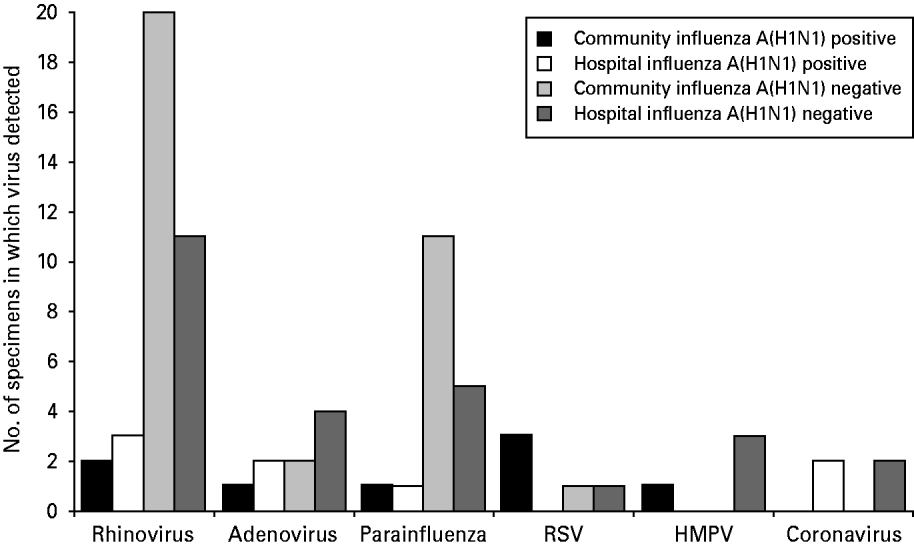
Fig. 1. Number of specimens positive for non-influenza respiratory viruses in each study group.
Of community specimens negative for influenza A(H1N1), 33·3% (31/93) were positive for at least one respiratory virus. Of these specimens, 29 were positive for one respiratory virus and two were positive for two viruses (parainfluenza with rhinovirus and parainfluenza with adenovirus).
Of the specimens from hospitalized patients negative for influenza A(H1N1), 30·0% (24/80) were positive for at least one respiratory virus. Of these specimens, 22 were positive for one respiratory virus and two were positive for two viruses (adenovirus with HMPV and parainfluenza with coronavirus). There was no significant association between detection of another respiratory virus in samples negative for influenza A and whether they were from community or hospitalized patients (P=0·74, Fisher's exact test).
The total numbers of specimens in each study group in which non-influenza respiratory viruses were detected are shown in Figure 1.
Rhinoviruses were the most common non-influenza viruses detected, accounting for 47·4% (36/76) of detections. All other viruses were much less frequently detected. Total numbers of virus detections were too small to draw any conclusions about the significance of differences in detection rates between the four study groups.
Specimen request forms were retrieved for all the community specimens negative for influenza A(H1N1) but positive for another respiratory virus. Eighteen out of 31 request forms gave no clinical information. The other 13 reported ‘flu-like’ symptoms. Of the 13 forms reporting symptoms five stated that the antiviral drug oseltamivir had been prescribed. Four of these five were positive for rhinovirus and one was positive for adenovirus.
DISCUSSION
By the end of the first wave of the influenza A(H1N1) pandemic in September 2009 it was estimated that there had been 144 000–670 000 cases in England with 12% of these cases from the West Midlands Region [3]. The large number of respiratory specimens we received from both community and hospital locations provided a rare opportunity to investigate respiratory virus co-infections during the influenza A(H1N1) outbreak and to assess whether these co-infections played any role in the severity of illness and in the hospitalization of patients.
Although the four groups of specimens selected were intended to be comparable with regards to age distribution, the influenza A(H1N1) positive specimens did cluster more closely to a mean age of ~16 years (Table 1). This age distribution of positives was a characteristic feature of the influenza A(H1N1) outbreak [1, Reference Scriven4, Reference Falagas18] which also explains the paucity of specimens available for study in the ⩾45 years age group.
In the UK, the majority of infected patients had self-limiting uncomplicated influenza with only 1·3–2·5% being hospitalized [3]. In this study, no statistically significant difference in the rate of respiratory virus co-infection in influenza A(H1N1)-positive patients was found between hospitalized or community patients. Although the numbers are small, this suggests that having another respiratory virus in addition to influenza A(H1N1) was not more likely to result in hospitalization, suggesting that hospitalization is determined by other underlying factors (such as chronic pulmonary or cardiac disease, obesity and pregnancy), as observed in other studies particularly in patients needing intensive-care facilities [Reference Scriven4, Reference Falagas18, 19].
One prominent feature of the results was the prevalence of rhinovirus compared to other non-influenza viruses. In the influenza A(H1N1)-negative community group 21·5% (20/93) of the specimens were positive for rhinovirus. If the study sample is representative of all the specimens received during the summer outbreak this could be extrapolated to estimate that about 290 of the 1361 community patients testing negative for influenza A(H1N1) could have had a rhinovirus infection. Considering that at least five influenza A(H1N1)-negative community patients with ‘flu-like’ symptoms due to rhinovirus or adenovirus were prescribed oseltamivir, it is likely that quite a number of influenza-negative patients with rhinovirus may have been prescribed oseltamivir.
Infections with the new influenza A(H1N1) generally produced mild, afebrile symptoms in most cases [Reference Cao20]. Characterization of the virus had shown that it lacked virulence factors associated with highly pathogenic influenza viruses [Reference Herfst21]. The observation that rhinovirus and adenovirus infections can be mistaken for influenza corroborates the known symptom overlap between influenza and other respiratory viruses [Reference Heikkinen and Jarvinen7]. Based on these observations it also seems possible that the National Pandemic Flu Service's helpline and website (running from July 2009 to February 2010) which allowed people with flu-like symptoms to obtain oseltamivir without presenting to healthcare facilities may have resulted in some inappropriate medication.
It is assumed that all specimens in this study were collected from suspected, symptomatic influenza A(H1N1) cases. The numbers of rhinovirus may indicate that it was the most commonly circulating non-influenza respiratory virus in the Midlands during the study period. However, it is also possible that rhinovirus was producing the most ‘flu-like’ symptoms prompting more testing for influenza A(H1N1). The clinical diagnosis of influenza, even in a pandemic, can be unreliable and require confirmation by virological assays.
ACKNOWLEDGEMENTS
Funding was received from the Health Protection Agency.
DECLARATION OF INTEREST
None.






