INTRODUCTION
Streptococcus pyogenes or group A streptococcus (GAS) is the aetiological agent of various exclusive human diseases spanning non-invasive ‘strep throat’ to necrotizing fasciitis or streptococcal toxic shock syndrome [Reference Carapetis1, Reference Baxter and McChesney2]. GAS is the most common bacterial cause of acute pharyngo-tonsillitis. Moreover, infection with S. pyogenes may lead to the development of auto-immune disorders such as rheumatic fever and associated heart disease and acute glomerulonephritis. During the 1980s and 1990s, outbreaks of streptococcal rheumatic fever and systemic disease with severe complications were registered in many countries worldwide [Reference Baxter and McChesney2–Reference Berrios4] and it has been suggested that this epidemiological change could have been due to an increase in the frequency of GAS types associated with severe infections [Reference Schwartz, Facklam and Breiman5], or an increase in strains with a high content of capsular hyaluronic acid (HA) [Reference Kaplan, Johnson and Cleary6]. The incidence of severe GAS diseases varies with time and geographical region, probably reflecting the susceptibility of the population to strains with particular virulence capabilities. GAS virulence depends on a variety of secreted and surface proteins that promote host invasion as well as evasion of the immune response. The HA capsule confers invasiveness in vivo, by interfering with the binding of antibodies leading to phagocytosis resistance [Reference Stollerman and Dale7]. Strains rich in HA are mucoid on agar culture and have been implicated in severe invasive infections [Reference Schrager, Rheinwald and Wessels8]. The synthesis of HA for capsule formation is controlled by an operon which is negatively regulated by a protein, CsrR, which binds upstream of the coding region of capsule biosynthetic genes and represses their transcription [Reference Dalton, Hobb and Scott9, Reference Heath10]. It has been shown that when this repressor is inactivated, HA is highly expressed and the infection produced is more invasive and severe [Reference Engleberg11]. M protein also confers resistance to phagocytosis as M-negative GAS strains are killed by this process [Reference Fischetti, Hodges and Hruby12]. M protein is encoded by the emm gene and to date more than 150 different emm gene types have been defined by sequence typing which due to its unambiguity and accuracy is considered the ‘gold standard’ for type identification of GAS [Reference Efstratiou13].
M-type distribution varies across geographical regions and individual countries [14, Reference Richardson15]. Previous studies of genetic diversity of GAS in Chile have been limited to a reduced number of strains with specific traits and their HA content was not addressed. We aimed to determine the emm types and the amount of HA present in GAS strains isolated in Chile from patients with invasive (i.e. recovery of GAS from sterile sites) and non-invasive (pharyngitis) infections over a 12-year period, and to assess the relationship between emm type and phenotypic expression of HA with regard to virulence.
METHODS
Bacterial strains
GAS clinical isolates from multiple health centres in Chile were routinely analysed in the Clinical Microbiology Laboratory of the Pontificia Universidad Católica de Chile and stored since 1996. Non-invasive isolates were recovered mainly from throat swabs of patients with pharyngitis and invasive isolates were from diverse sterile sites of infected patients: blood, synovial fluid, bronchial secretion, abscesses and biopsy tissue. From a total of 1282 isolates from the period 1996–2007, 110 (30 invasive, 80 non-invasive) were randomly selected using computer software (Minitab) for the current study. Isolates were stored in brain-heart infusion broth (Merck, Germany) supplemented with 5% sheep blood and subcultured on Columbia blood agar (bioMérieux, France).
emm gene typing
Strains were sequenced according to Centers for Disease Control and Prevention (CDC) protocol [16]. Briefly, DNA was extracted from the isolates using the Qiagen DNA extraction kit (Qiagen, Germany), amplified by PCR with standard primers [Reference Whatmore17] and following column purification were sequenced using the M1a primer [Reference Whatmore17]. The sequence obtained was compared with the CDC database and assigned an emm type [16]. In accordance with CDC criteria, a new emm type was defined if a sequence identity of <92% with the reference type (identified as subtype .0) was observed in the 90 nucleotides encoding the first 30 amino acids of the processed M protein. Subtypes were assigned based on any change in the sequence of the first 50 amino acids of the processed M protein. All sequences found to be different from the reference sequence were searched for in the GenBank database using the BLAST program to confirm their novelty.
csrR gene sequencing
A product of 714 bp was amplified with the following primers: CsrF: GGGTTGGTATAAATGACAAAG and CsrR: GATTTTCCATATGACTTATTTC and this was sequenced with CsrF primer. Sequences were compared to a wild-type csrR gene sequence (accession no. AB513958.1) using the ClustalW programme. The dendrogram was produced using UPGMA cluster analysis [Reference Sneath and Sokal18] and the maximum composite likelihood method [Reference Tamura, Nei and Kumar19] with Mega4 software [Reference Tamura20]. All sequences that showed variations with respect to the wild-type sequence were searched for in the GenBank database to confirm their novelty.
Mucoid phenotype and capsular HA content
The expression of capsule by isolates was determined by visual inspection of colonies grown on Columbia blood agar for 24 h with 5% of CO2. Mucoid (capsulate) isolates were characterized by a brilliant halo while non-mucoid strains grew with a matt appearance. For the quantification of capsular HA the Stains-all method was used [Reference Darmstadt21]. Briefly, isolates were cultivated for about 4–6 h in Todd–Hewitt broth (Becton-Dickinson, France) at 37°C to an optical density (600 nm) of 0·6–0·8. After plating serial dilutions of this broth on Columbia blood agar for colony-forming unit (c.f.u.) quantification, bacteria were centrifuged at 8000 g, washed with distilled water and incubated at room temperature with chloroform for 1 h to release capsular HA. After centrifugation at 12 400 g for 5 min, 100 μl of the supernatant were added to 1 ml of freshly prepared Stains-all reagent (Sigma, USA) and the absorbance was measured at 640 nm. To quantify HA content, a calibration curve was prepared with known concentrations of a reference HA (EMD Biosciences, USA) (0·005, 0·01, 0·02, 0·03 μg/μl). The amount of HA was expressed as femtograms per c.f.u. (fg/c.f.u.). Reference control strains of mucoid (87–282 [Reference Ashbaugh, Alberti and Wessels22]) and non-mucoid (771cap− [Reference Schrager, Rheinwald and Wessels8]) were kindly provided by Dr Michael Wessels (Medical School, Harvard University, USA).
Erythromycin susceptibility testing
Erythromycin susceptibility was determined for all isolates by the agar diffusion method according to Clinical and Laboratory Standards Institute guidelines (CLSI, 2006) [23]. Reference strains of known antibiotic susceptibility were used as controls and inhibition zone diameters were measured and interpreted according to CLSI criteria [24].
Statistical analysis
One-way analysis of variance (ANOVA), Student's unpaired two-tailed t test and χ2 analysis were performed with GraphPad Prism 5.0 software (GraphPad, USA). To measure the association between csrR gene and emm type, 2×2 contingency tables were used. GraphPad software was used to calculate statistical significance using χ2 analysis with Yates' correction for low sample size.
RESULTS
emm-type distribution among GAS clinical isolates
Nineteen different emm types were identified and the most frequent were emm12 (19·1%), emm1 (14·5%), emm4 (11·8%), and emm28 (8·2%); the six predominant emm types accounted for 67% of the total isolates (Table 1). Sixteen different emm types were found among pharyngeal isolates, 10 of which were absent from sterile-site isolates (emm4, emm2, emm77, emm9, emm94, emm78, emm87, emm3, emm73, emm89). By contrast, the 30 sterile-site isolates were represented by nine different emm types, three of which (emm11, emm48, emm81) were absent from pharyngeal isolates. Type emm4 was frequent among pharyngeal isolates but was not found in sterile sites. Similarly, type emm75 accounted for 13·3% of sterile-sites isolates but only 2·5% of pharyngeal isolates.
Table 1. emm-type distribution in invasive and non-invasive clinical isolates in Chile, 1996–2007
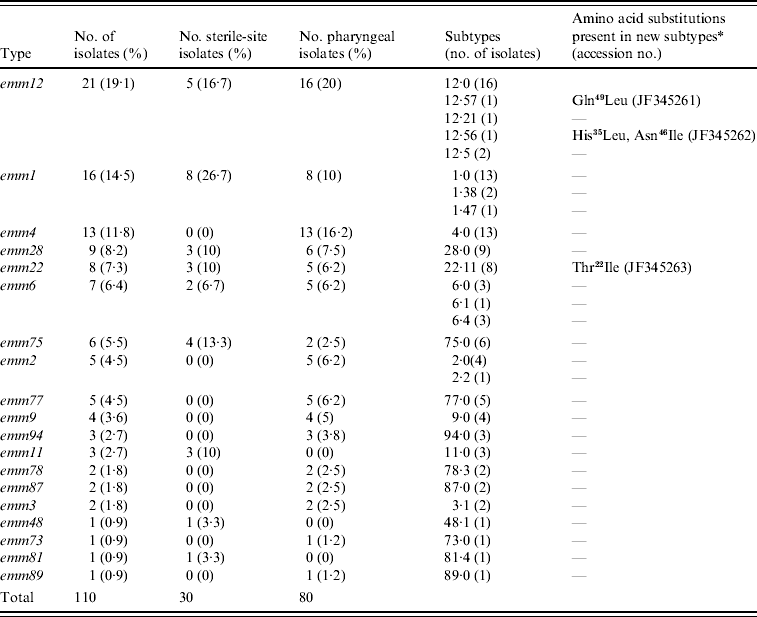
* Differences with respect to the reference subtype sequence of the CDC database. Amino acid positions correspond to the first 50 amino acids of the mature protein. Gln49Leu, glutamine at position 49 changed to leucine; His35Leu, histidine at position 35 changed to leucine; Asn46Ile, asparagine at position 46 changed to isoleucine; Thr22Ile, threonine at position 22 changed to isoleucine.
Of the 19 emm types identified, all but five contained representatives of the reference type (.0) but four types (emm1, emm2, emm6, emm12) were heterogeneous in subtype (Table 1); emm12 comprised five subtypes with the reference type 12·0 being the most predominant, in contrast with emm2 which had two subtypes represented by four and one isolates. The emm sequences of three isolates were novel with reference to CDC and GenBank databases and therefore constitute new emm subtypes. These were deposited in the GenBank database with accession numbers JF345261, JF345262 and JF345263 and in the CDC database as new emm subtypes. Their new designations and specific changes in amino acid sequence are given in Table 1.
Figure 1 shows the temporal distribution of emm types over the study period. Half of the 110 isolates were recovered during 2000–2002 (17, 19, 19 isolates, respectively) and these exhibited the greatest diversity in type distribution. Type emm1 was found in all but two years, whereas emm12 and emm4 emerged later in 2000 and 2001, respectively, and emm22 disappeared in later years.
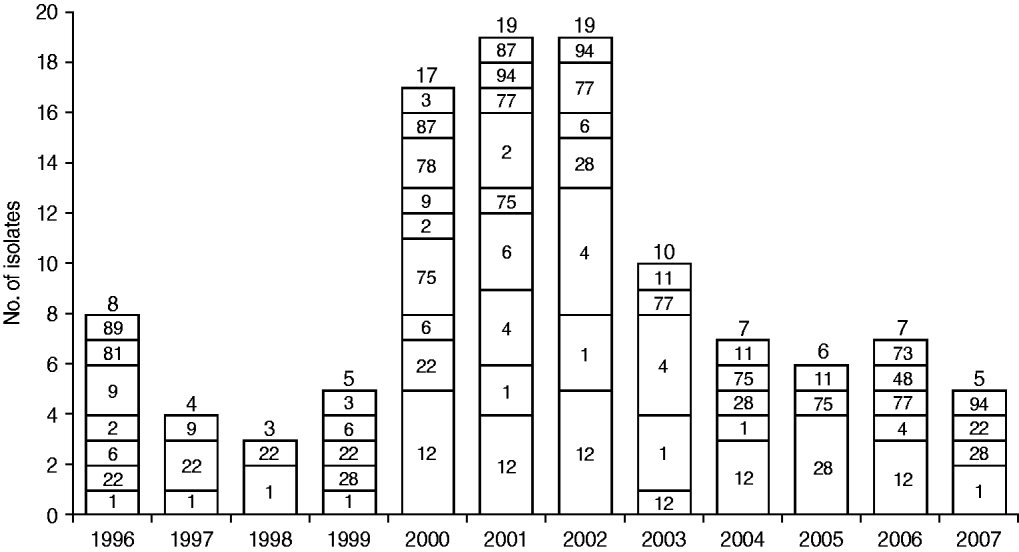
Fig. 1. Temporal distribution of emm types in the group A streptococcus population. The number of isolates from each year is shown above each bar and the corresponding emm types are shown inside bars.
Mucoid phenotype, HA content and correlation with infection site and emm type
Inspection of colony morphology showed that three isolates (nos. 25, 128, 147) were mucoid in appearance. The HA content of all 110 isolates was quantified and capsulate strains were distinguished from non-capsulate strains by HA values of >10 fg/c.f.u [Reference Schrager, Rheinwald and Wessels8, Reference Jadoun, Eyal and Sela25]. Mucoid isolate nos. 25 and 128 had an HA content of 11·8 and 23·4 fg/c.f.u, respectively, but isolate no. 147 had only 8·4 fg/c.f.u in spite of its mucoid appearance. The latter isolate was from a sterile site and type emm1 and the former two were from the pharynx and emm12. The HA content for the remainder of the isolates ranged between 0·2 and 8·4 fg/c.f.u with significantly higher values observed for sterile-site isolates compared to pharyngeal isolates (P=0·0374) (Fig. 2a). There was no statistical difference between HA content and emm type by ANOVA analysis (P=0·9128) (Fig. 2b). Forty-six (41·7%) isolates were resistant to erythromycin and almost half of these were emm4 and emm12 (Fig. 2c). Erythromycin-resistant isolates had significantly lower HA content than susceptible isolates (Fig. 2d).
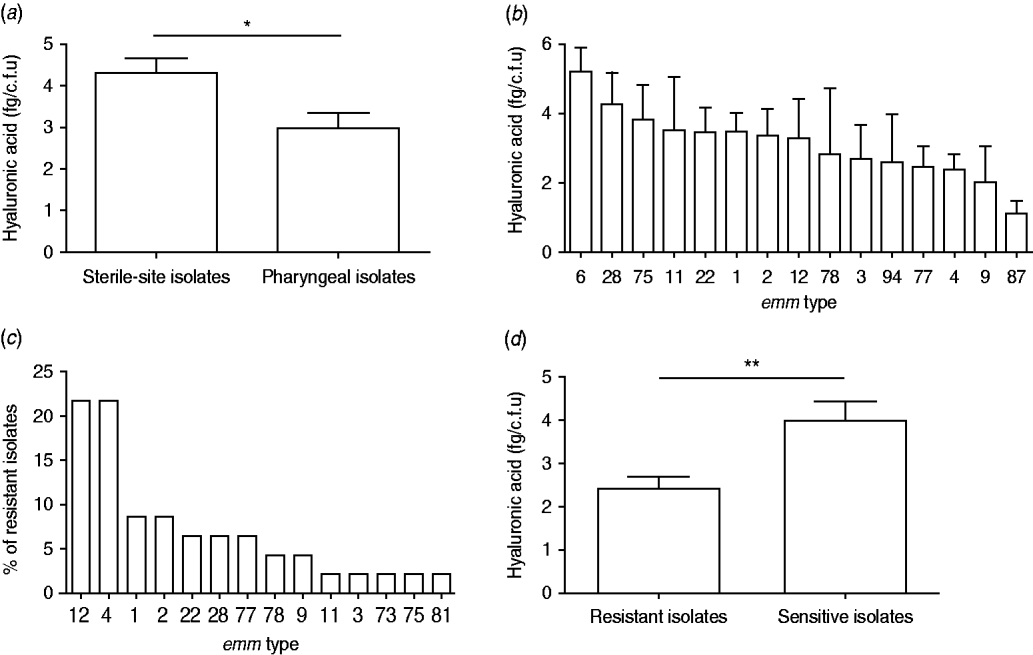
Fig. 2. Hyaluronic acid (HA) content, emm type and erythromycin resistance of 110 group A streptococcus clinical isolates. HA content (fg/c.f.u) of (a) pharyngeal and sterile-site isolates and (b) most prevalent emm types. (c) emm-type distribution among 46 erythromycin-resistant isolates, and (d) HA content of erythromycin-resistant and susceptible isolates. Data are means of two or three independent experiments and bars represent the standard error (Student's t test: * P<0·05, ** P<0·005).
Sequence analysis of the csrR gene
The coding region of the csrR gene of all isolates was sequenced to determine if the increased expression of HA of the culturally mucoid strains was due to point mutations, i.e. single nucleotide polymorphisms (SNPs), in this gene. The majority (77/110) of isolates had SNPs in the csrR gene; they had combinations of 1–4 SNPs. Each combination was defined as an allelic type (AT) (Table 2). Most SNPs did not result in an amino acid substitution but four of them did produce amino acid substitutions: methionine-170 for isoleucine (Met170Ile), valine-173 for alanine (Val173Ala), arginine-94 for cysteine (Arg94Cys) and glycine-91 for aspartic acid (Gly91Asp) (Table 2). Amino acid substitutions Met170Ile and Val173Ala were located in the DNA-binding domain and found in non-mucoid isolates only whereas Arg94Cys and Gly91Asp were located in the active site of the regulator and were present only in mucoid isolates nos. 25 and 128, respectively; no mutation was identified in mucoid isolate no. 147. The novel csrR gene sequences were deposited in the GenBank database with accession numbers JF345257, JF345258, JF345259 and JF345260.
Table 2. Allele types of csrR gene found in the group A streptococcus population
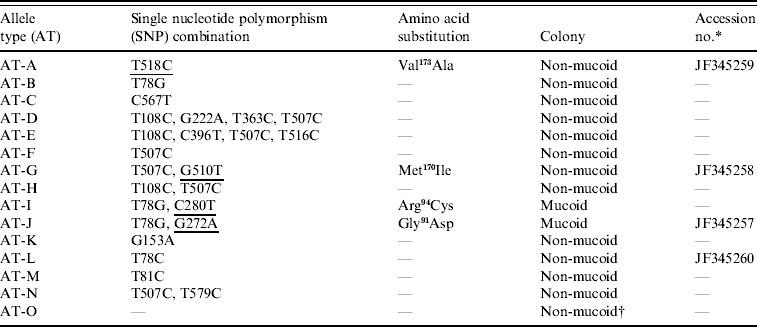
The SNPs producing amino acid substitutions are underlined.
* Accession no. is given for the novel sequences found in this work.
† Isolate no. 147 was the only mucoid strain of the type AT-O.
Association of emm type with csrR allele types
A dendrogram was constructed based on the csrR gene sequences to analyse the AT distribution of csrR gene in the population and its association with emm type (Fig. 3). Most ATs were heterogeneous and not associated with a particular emm type as exemplified by the most frequent AT-O, where 11 different emm types were identified, although 36% were of type emm1. However, it should be noted that all AT-L and AT-M isolates were type emm28 and all AT-E isolates were type emm6; other associations were 62% of AT-C isolates and 75% of AT-D isolates were of types emm12 and emm4, respectively. In order to statistically ascertain the association between ATs of csrR gene and emm type, ATs were chosen as the genotypically distinct units for comparison and the emm type as the variable unit. Pairwise associations between ATs represented by more than five isolates (AT-O, AT-C, AT-F, AT-D, AT-G) and the more represented emm types of these ATs, revealed statistically significant levels of non-random association between AT-O and emm1, AT-C and emm12, AT-F and emm22, AT-D and emm9, and AT-G and emm77 (Table 3). Type emm12 was significantly associated with AT-C compared to other ATs, emm4 with AT-D, and emm77 with AT-G. However, type emm1 was significantly associated with AT-O compared to AT-C and AT-D and similarly, emm22 with AT-F compared to AT-C and AT-O (Table 3).
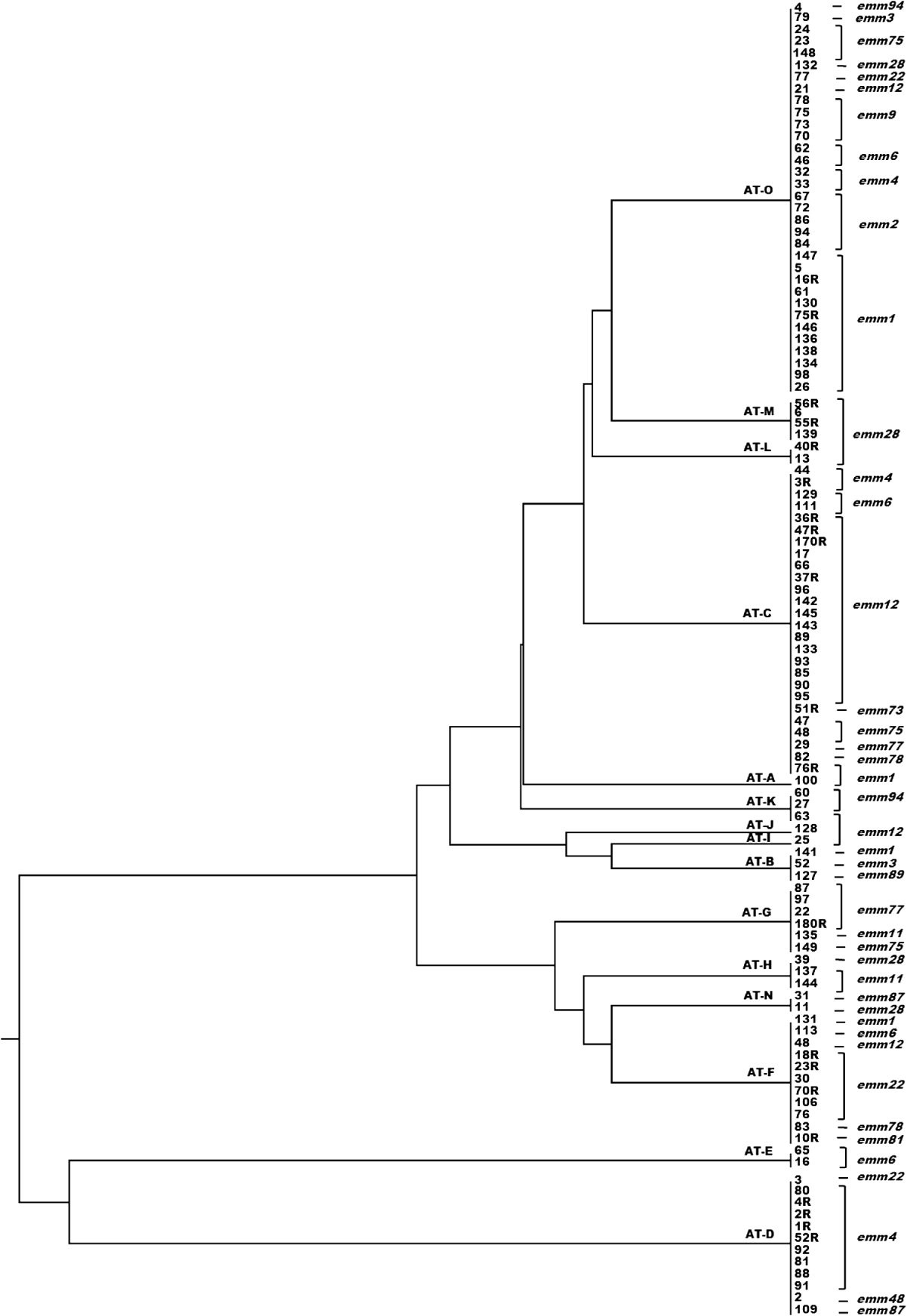
Fig. 3. Dendrogram from UPGMA cluster analysis of the csrR gene sequence of 110 group A streptococcus isolates showing allele type (AT) and emm type.
Table 3. Statistical analysis of allele-type and emm-type associations in isolates of distinct allele types
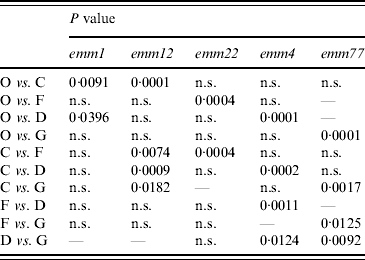
χ2 analysis (two-tailed) with Yates' correction for sample size. P<0·05 indicates that null hypothesis for random assortment is rejected.
P>0·05, not significant (n.s.)
DISCUSSION
This is the first report of emm types and HA content of GAS strains in Chile determined in a population of more than 100 strains isolated over more than a decade. There are temporal variations in the emm-type distribution which may be due to epidemiological changes of GAS. In our study, nine of the sterile-sites isolates came from blood cultures and four (44%) of them were of type emm1 in accordance with published reports [Reference Schwartz, Facklam and Breiman5, Reference Johnson, Stevens and Kaplan26]. The data presented here indicate that emm-type distribution in Chile is similar to that reported for Latin America and developed countries [14]. According to data reported by Espinosa et al. [Reference Espinosa27], nine of the ten most frequent emm types in Chile and Mexico are common to both countries. However, a similar comparison with India shows only three emm types in common [Reference Sagar28]. It appears that emm-type distribution among invasive isolates is more conserved in different countries as the most frequent types of invasive infections in Argentina [Reference Lopardo29], USA [Reference O'Loughlin30], Mexico [Reference Espinosa27] and Italy [Reference Creti31] are emm1 and emm12, the same as in Chile. The oligoclonal distribution of emm types in Chile is similar to that reported from Belgium [Reference Smeesters32] and the USA [Reference O'Loughlin30] but in contrast with that reported from Brasilia (Brazil) where the most abundant emm type had a frequency of only 8·5% which is more indicative of a polyclonal distribution [Reference Smeesters32] as also found in India and Lebanon [Reference Sagar28, Reference Bahnan33]. emm-type distribution possibly reflects the type of population of a country or region and those regions which have experienced extensive recent migrations increasing host population diversity thus have a greater diversity of emm types. This could also be due to ethnic diversity of geographical regions. The low diversity of emm types in Chile may therefore reflect the homogeneous nature and stability of its population.
The average HA content did not vary significantly by emm type, possibly because differences were not detectable due to the low number of isolates of some emm types. However, we found a significant correlation between increased amount of capsular HA and sterile sites of isolation (invasiveness). This issue has been addressed by Schrager et al. [Reference Schrager, Rheinwald and Wessels8] who suggested that HA capsule was a virulence factor in soft-tissue infections as capsulate strains avoid uptake by epithelial cells in contrast to unencapsulated ones and this could favour entrance through an extracellular route leading to tissue necrosis and spread-through infection. The analysis of mutations in CsrR repressor showed that mutations Met170Ile and Val173Ala are located in the C-terminal region of the CsrR repressor involved in DNA binding. Isolates bearing these mutations are non-mucoid perhaps because both amino acids are not directly involved in DNA binding [Reference Miyoshi-Akiyama34]. Mutations Arg94Cys and Gly91Asp are located in the active site of the regulator and both of these are associated with increased amount of HA and mucoid colony phenotype. It has been reported that mutations in this region of the protein confer mucoid character [Reference Engleberg11]. However, mucoid isolate no. 147 described here has no mutations in its csrR gene which suggests that factors other than repressor inactivation might contribute to increased capsule synthesis.
M protein is a target of the protective immune response, and amino acid substitutions in this protein would allow the pathogen to evade the immune response. The emm gene is therefore subjected to a high selective pressure and may change rapidly resulting in emm-type variation among isolates. By contrast, housekeeping genes are not subject to the same pressure and change at a much lower rate. The csrR gene, although not a housekeeping gene, encodes a global regulator which is not expected to be under high selective pressure as the emm gene. Therefore, the association between emm type and csrR sequence is a valid means of estimating horizontal gene transfer in the GAS population. Genetic linkage between emm gene markers and spe alleles have been described in GAS isolates [Reference Bessen35] but to our knowledge this is the first report of a correlation existing between emm and csrR genes. The non-random association of csrR alleles with emm types could be due to the strong clonality of GAS that causes csrR and emm genes to change together. These observations support the concept that free horizontal gene transfer is hindered by certain barriers, otherwise many random combinations of ATs and emm types would be observed. This picture is compatible with an oligoclonal GAS population, as observed in Chile, in which there is a lack of incoming new strains due to its geographical and social characteristics. The geographical isolation of Chile is supported by the absence of both community-acquired methicillin-resistant Staphylococcus aureus and carbapenemase-producing Enterobacteriaceae which are very frequent in other South American countries including Argentina [Reference Radice36, Reference Tokumoto37], Uruguay [Reference Benoit38], Colombia [Reference Alvarez39, Reference Villegas40] and Brazil [Reference Peirano41, Reference Ribeiro42].
The most recent published study of emm-type distribution in Chile in 2001 [Reference Palavecino43], addressed antimicrobial-resistant isolates only. Our data suggest that the emm-type distribution among erythromycin-resistant isolates has changed since then as type emm2, which represented 73% of resistant isolates in the period 1994–1998 has decreased to 7% in the present study. This could be due to the decrease of erythromycin resistance observed in recent years [Reference Rodríguez44].
There is no GAS vaccine yet available although these organisms cause more deaths than, e.g. Neisseria meningitidis, against which a vaccine is available. A multivalent vaccine based on purified M-protein peptides of the 26 most prevalent emm types is currently undergoing the Phase II stage of development in the USA [Reference Hu45]. If the US vaccine was used in Chile, it would protect against only 58% of the most frequent emm types found in this study. It is necessary to know the emm types that are present in the population in order to allow the evaluation of the effectiveness of future GAS vaccines in this population. Continued surveillance of emm-type distribution among invasive and non-invasive GAS is therefore required to guide the design and use of potential vaccines.
ACKNOWLEDGEMENTS
This work was supported by the Department of Clinical Laboratories at Pontificia Universidad Católica de Chile, the Sentry Programme, and Millennium Nucleus of Immunology and Immunotheraphy. We thank Dr Guido Mora for critical reading of the manuscript and Cecilia Zumarán and Pablo Rojas Zuñiga for technical assistance. We also express our gratitude to Bernard Beall and Yusra Ahmad for helping with emm sequence typing interpretation, and to the CDC database for new emm sequence subtype assignment.
DECLARATION OF INTEREST
None.










