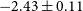36 results
A monitoring campaign (2013–2020) of ESA’s Mars Express to study interplanetary plasma scintillation
-
- Journal:
- Publications of the Astronomical Society of Australia / Volume 40 / 2023
- Published online by Cambridge University Press:
- 12 April 2023, e013
-
- Article
-
- You have access
- Open access
- HTML
- Export citation
Visualising the proximal urethra by MRI voiding scan: results of a prospective clinical trial evaluating a novel approach to radiotherapy simulation for prostate cancer
-
- Journal:
- Journal of Radiotherapy in Practice / Volume 21 / Issue 4 / December 2022
- Published online by Cambridge University Press:
- 05 April 2021, pp. 472-475
-
- Article
-
- You have access
- Open access
- HTML
- Export citation
EPA-0229 - Association Between Depression and Mortality Depends on Methodology Used
-
- Journal:
- European Psychiatry / Volume 29 / Issue S1 / 2014
- Published online by Cambridge University Press:
- 15 April 2020, p. 1
-
- Article
-
- You have access
- Export citation
Simultaneous temporal trends in dementia incidence and prevalence, 2005–2013: a population-based retrospective cohort study in Saskatchewan, Canada
-
- Journal:
- International Psychogeriatrics / Volume 28 / Issue 10 / October 2016
- Published online by Cambridge University Press:
- 29 June 2016, pp. 1643-1658
-
- Article
-
- You have access
- Open access
- HTML
- Export citation
Association between vitamin C status and CVD and mortality risk in France and Northern Ireland: the PRIME study
-
- Journal:
- Proceedings of the Nutrition Society / Volume 73 / Issue OCE2 / 2014
- Published online by Cambridge University Press:
- 24 September 2014, E80
-
- Article
-
- You have access
- HTML
- Export citation
Contributor affiliations
-
-
- Book:
- Textbook of Neural Repair and Rehabilitation
- Published online:
- 05 May 2014
- Print publication:
- 24 April 2014, pp ix-xvi
-
- Chapter
- Export citation
Contributor affiliations
-
-
- Book:
- Textbook of Neural Repair and Rehabilitation
- Published online:
- 05 June 2014
- Print publication:
- 24 April 2014, pp ix-xvi
-
- Chapter
- Export citation
Association between diet and cardiovascular disease and overall mortality risk in France and Northern Ireland: the PRIME study
-
- Journal:
- Proceedings of the Nutrition Society / Volume 72 / Issue OCE3 / 2013
- Published online by Cambridge University Press:
- 28 August 2013, E160
-
- Article
-
- You have access
- HTML
- Export citation
Breeding for robust cows that produce healthier milk: RobustMilk
-
- Journal:
- Advances in Animal Biosciences / Volume 4 / Issue 3 / July 2013
- Published online by Cambridge University Press:
- 30 July 2013, pp. 594-599
- Print publication:
- July 2013
-
- Article
- Export citation
A comparison of dietary patterns of middle aged men in France and Northern Ireland: the PRIME Study
-
- Journal:
- Proceedings of the Nutrition Society / Volume 71 / Issue OCE2 / 2012
- Published online by Cambridge University Press:
- 19 October 2012, E102
-
- Article
-
- You have access
- HTML
- Export citation
Frontmatter
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp i-iv
-
- Chapter
- Export citation
Appendix A - Global optimization algorithms
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp 262-276
-
- Chapter
- Export citation
Index
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp 281-282
-
- Chapter
- Export citation
Acknowledgements
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp xi-xii
-
- Chapter
- Export citation
Contents
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp v-viii
-
- Chapter
- Export citation
About the authors
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp 277-280
-
- Chapter
- Export citation
Preface
-
- Book:
- Optimal Device Design
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009, pp ix-x
-
- Chapter
- Export citation

Optimal Device Design
-
- Published online:
- 04 May 2010
- Print publication:
- 24 December 2009