Article contents
Synchrotron powder diffraction, X-ray absorption and 1H nuclear magnetic resonance data for hypoxanthine, C5H4N4O
Published online by Cambridge University Press: 13 August 2015
Abstract
Synchrotron powder X-ray diffraction, X-ray absorption spectroscopy (XAS), and proton nuclear magnetic resonance (1H-NMR) data have been used to examine the structure of hypoxanthine, 1,7-dihydro-6H-purin-6-one (C5H4N4O), a purine base that participates in numerous metabolic processes. XAS and 1H-NMR spectroscopy were used to determine that hypoxanthine was present in its keto form (both in solid state and dissolved in an organic solvent). Rigid body refinement was performed with the Rietveld software package GSAS yielding triclinic lattice parameters of a = 7.1179 (2) Å, b = 9.7830 (3) Å, c = 10.4009 (3) Å, α = 58.876 (1)°, β = 67.609 (1)°, and γ = 71.937 (2)° (C5H4N4O, Z = 4, space group P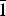 $\bar 1$).
$\bar 1$).
Keywords
- Type
- New Diffraction Data
- Information
- Copyright
- Copyright © International Centre for Diffraction Data 2015
References
- 8
- Cited by




