Article contents
Fast Improvement of TEM Images with Low-Dose Electrons by Deep Learning
Published online by Cambridge University Press: 10 December 2021
Abstract
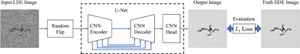
Low electron dose observation is indispensable for observing various samples using a transmission electron microscope; consequently, image processing has been used to improve transmission electron microscopy (TEM) images. To apply such image processing to in situ observations, we here apply a convolutional neural network to TEM imaging. Using a dataset that includes short-exposure images and long-exposure images, we develop a pipeline for processed short-exposure images, based on end-to-end training. The quality of images acquired with a total dose of approximately  $5$
$5$  $e^{-}$ per pixel becomes comparable to that of images acquired with a total dose of approximately
$e^{-}$ per pixel becomes comparable to that of images acquired with a total dose of approximately  $1{,}000$
$1{,}000$  $e^{-}$ per pixel. Because the conversion time is approximately 8 ms, in situ observation at 125 fps is possible. This imaging technique enables in situ observation of electron-beam-sensitive specimens.
$e^{-}$ per pixel. Because the conversion time is approximately 8 ms, in situ observation at 125 fps is possible. This imaging technique enables in situ observation of electron-beam-sensitive specimens.
- Type
- Software and Instrumentation
- Information
- Copyright
- Copyright © The Author(s), 2021. Published by Cambridge University Press on behalf of the Microscopy Society of America
References
- 10
- Cited by





