No CrossRef data available.
Article contents
Automated ExEm-spFRET Microscope
Published online by Cambridge University Press: 21 February 2022
Abstract
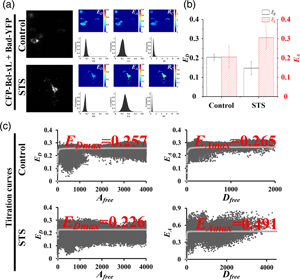
Excitation–emission-spectral unmixing-based fluorescence resonance energy transfer (ExEm-spFRET) microscopy exhibits excellent robustness in living cells. We here develop an automatic ExEm-spFRET microscope with 3.04 s of time resolution for a quantitative FRET imaging. The user-friendly interface software has been designed to operate in two modes: administrator and user. Automatic background recognition, subtraction, and cell segmentation were integrated into the software, which enables FRET calibration or measurement in a one-click operation manner. In administrator mode, both correction factors and spectral fingerprints are only calibrated periodically for a stable system. In user mode, quantitative ExEm-spFRET imaging is directly implemented for FRET samples. We implemented quantitative ExEm-spFRET imaging for living cells expressing different tandem constructs (C80Y, C40Y, C10Y, and C4Y, respectively) and obtained consistent results for at least 3 months, demonstrating the stability of our microscope. Next, we investigated Bcl-xL-Bad interaction by using ExEm-spFRET imaging and FRET two-hybrid assay and found that the Bcl-xL-Bad complexes exist mainly in Bad-Bcl-xL trimers in healthy cells and Bad-Bcl-xL2 trimers in apoptotic cells. We also performed time-lapse FRET imaging on our system for living cells expressing Yellow Cameleon 3.6 (YC3.6) to monitor ionomycin-induced rapid extracellular Ca2+ influx with a time interval of 5 s for total 250 s.
Keywords
- Type
- Software and Instrumentation
- Information
- Copyright
- Copyright © The Author(s), 2022. Published by Cambridge University Press on behalf of the Microscopy Society of America





