INTRODUCTION
Emerging infectious respiratory diseases constitute a continuing public health threat worldwide. Rapid movement of people and goods around the world increases the opportunity for local outbreaks to quickly become worldwide pandemics. The 2009 H1N1 swine influenza pandemic [Reference Garten1, 2] and continuing circulation and evolution of H5N1 avian influenza isolated from humans [Reference Buchy3, Reference Webster and Govorkova4] underscores a need for safe specimen collection kits that: (1) are compatible with molecular diagnostic and genomic analysis, (2) can effectively inactivate potentially harmful microbes, (3) stabilize and preserve nucleic acids including RNA, and (4) can be easily used in the field and transported at ambient temperatures while preserving microbial RNA.
Until recently, the majority of clinical diagnostic laboratories employed traditional culture for pathogen identification that typically requires 3–7 days for most viruses [Reference Daum5] and longer for some bacterial strains [Reference Zoetendal, Vaughan and de Vos6]. Traditional culture requires specimen collection of viable microbes, frozen transport, propagation and handling of potentially infectious and often unknown biological microbes. Furthermore, many infectious agents, e.g. highly pathogenic avian influenza, SARS, etc., are BSL-3 level pathogens that require specialized facilities and precautions for analysis. There are challenges in obtaining, shipping and maintaining high-quality, viable biological specimens for culture. Specimens must be shipped using a cold chain, most often dry ice. Transporting potentially infectious samples from remote sites or across international borders using commercial transit can be costly and tedious, particularly when specimens must be received frozen.
The field of clinical molecular diagnostics changed drastically with the advent of polymerase chain reaction (PCR) [Reference Saiki7], and subsequently, real-time PCR [Reference Higuchi8]. Real-time reverse transcription–PCR (rRT–PCR) can deliver superior sensitivity and specificity results in hours [Reference Daum9]. Thus, the majority of current diagnostic laboratories have transitioned from traditional culture to nucleic acid testing (NAT) such as real-time PCR.
Collection is the first step in diagnostic platforms or molecular protocols requiring the detection of potentially minute amounts of nucleic acids from microbes. Regardless of the nucleic acid test used or the RNA/DNA extraction protocol, specimen collection, specifically the inactivation of potentially infectious agents and the preservation and stability of pathogen RNA/DNA remains a critical gap in clinical diagnostics, especially for use around the world.
This work describes the use of PrimeStore Molecular Transport Medium (MTM; Longhorn Vaccines & Diagnostics, USA), a clinical or environmental sample collection system specifically formulated for downstream molecular diagnostic testing. PrimeStore MTM is an optimized blend of proprietary reagents for RNA preservation that also includes the first non-specific exogenous internal positive control (IPC) for tracking the integrity of a specimen from the point of collection to detection. PrimeStore MTM is shown to efficiently: (1) lyse/inactivate potentially infectious biological pathogens reducing infection risk so that samples and can be handled, shipped, or transported with minimal fear of pathogen release or contamination, (2) stabilize and protect lysed ‘naked’ RNA polymers from hydrolysis, oxidative damage or nuclease degradation, (3) preserve RNA for prolonged periods at ambient temperature until they can be processed using NAT, and (4) be compatible with commercially available bench-top extraction kits.
MATERIALS AND METHODS
Microbe killing
Membrane filtration technique for bacterial and fungal recovery
The membrane filtration method for bacterial and fungal recovery was used to assess the killing ability of PrimeStore MTM. Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus [non-methicillin-resistant Staphylococcus aureus (MRSA)], Candida albicans, Bacillus subtilis, and Aspergillus brasiliensis were used to determine whether PrimeStore MTM could effectively kill and inactivate a panel of bacteria and mould (yeast and filamentous fungi). Positive controls incubated in a water matrix were performed on day 0 only. A population of 1×106 c.f.u. for each bacterial strain was inoculated into 0·5 ml PrimeStore for each time-point and subsequently incubated at 20–25°C. The containers were enumerated and evaluated at days 0, 1, 7, 14 and 28. The inoculum was aseptically passed through a sterile filtration device and subsequently rinsed three times with 100 ml sterile neutralizing fluid D [1 g peptic digest of animal tissue (peptone) and 1 ml polysorbate 80 dissolved in 1·0 l of sterile water (final pH 7·1±0·2)]. Where necessary, dilutions of the inoculated test article were performed to deliver a target count of 25–250 c.f.u. per filter. For each time-point, inoculated negative controls were processed in a similar fashion. Filters inoculated with samples containing bacteria were plated onto tryptic soy agar (TSAP) with lecithin and polysorbate 80 and incubated at 30–35°C for 72 h. Filters inoculated with samples containing yeast or mould were plated onto Sabouraud dextrose agar (SAB) and incubated at 20–25°C for no less than 72 h but no more than 5 days. Colonies were counted to calculate log10 recoveries and percent (%) kill for each organism used during microbial challenge.
Viral killing
Equal volumes of A/Wuhan/359/95 (108 TCID50/ml) or adenovirus (type 5) (108 TCID50/ml) and PrimeStore MTM were incubated for 10-, 30-, and 60-s intervals at ambient temperature. Influenza virus only and PrimeStore MTM (minus virus) were also tested as controls. After incubation, each solution was subjected to a tenfold serial dilution, and inoculated onto a 96-well plate confluent with MDCK cells (influenza) or A549 (adenovirus). Prior to influenza virus or adenovirus inoculation, 96-well plates were washed 2× with EMEM or EMEM containing 1 μg/ml trypsin, respectively. All samples were processed in quadruplicate. Plates were incubated at 37°C with 5% CO2 for 4 days, stained with Crystal Violet reagent (0·06%) in 1% glutaraldehyde for 1 h and rinsed with sterile water. Cells not stained exhibited cytopathic effect (CPE) of virus. The titre of the virus was recorded as the TCID50, i.e. the inverse of the dilution that resulted in the CPE in 50% of the wells. Viral titres were calculated as geometric means+standard error.
For testing against influenza A/Vietnam/1203/04(H5N1) virus (107 TCID50/ml), a collection swab was loaded with 0·1 ml H5N1 virus and placed in 1·5 ml PrimeStore MTM. Positive controls consisted of virus-loaded swabs in the absence of PrimeStore MTM, and swabs in PrimeStore MTM without influenza virus were used as negative controls. After 60-min incubation at ambient temperature, the samples were vortexed and inoculated into 10-ml cell culture media. Samples were serially diluted (tenfold) and inoculated onto 96-well plates confluent with MDCK cells. Plates were incubated at 37°C with 5% CO2 for 4 days and assessed for viral CPE.
MRSA killing
A stock plate containing about 108 c.f.u. MRSA (ATCC 33592) was transferred to TSB, vortexed briefly and incubated at ambient temperature for 10 min. A total of 0·1 ml bacterial suspension was transferred to 0·9 ml PrimeStore and vortexed for 60 s. A total of 0·1 ml suspension was transferred to 0·3 ml TSB (1:4 dilution) and 100 μl was transferred to blood agar plates (5% sheep RBCs in TSA) after 0, 5 and 15 min. Positive controls included equivalent volumes of MRSA and TSB. Plates were allowed to dry, incubated overnight at 37°C and analysed for c.f.u./ml.
Nuclease digestion experiment
Stock influenza A(H3N2) virus (103 TCID50/ml) spiked into human nasal washing was preserved in PrimeStore MTM or other indicated media (0·3 ml solution to 0·1 ml virus) and incubated at 37°C with RNase A (25 U), RNase T1 (125 U) and Turbo DNase (2 U) for 1, 24 and 48 h (Life Technologies Inc., USA). Reactions were inactivated at 75°C for 2 min prior to RNA extraction.
Test samples and RNA extraction
For the RNA stabilization and preservation experiments described in Figures 2 and 3, a human throat swab containing a respiratory oral/pharyngeal matrix was placed into collection tubes containing 1·5 ml PrimeStore MTM and viral transport medium (VTM; BD Universal Viral Transport Medium; USA). An influenza culture containing 102 TCID50/ml (Fig. 2) or 104 TCID50/ml (Fig. 3) of A/California/04/2009 influenza A(H1N1) was spiked into each PrimeStore MTM or VTM tube and incubated at the indicated temperature (4, 25 or 37°C).
RNA was extracted using the Ambion RNaqueous-Micro kit (Life Technologies) according to the manufacturer's protocol. For PrimeStore compatibility experiment with commercial extraction kits (Fig. 4), the Ambion RNAqeuous-Micro (Ambion cat. no. 1931), QIAamp Viral RNA Mini kit (cat. no. 52904), Invitrogen Charge Switch Total RNA Cell kit (cat. no. CS14010), and Ambion MagMAX Viral RNA Isolation kit (cat no. 1928) were utilized according to the manufacturer's recommendations with 100 μl of 103 or 101 TCID50/ml A/California/04/2009 influenza A(H1N1) stock culture. All time-point extractions were frozen at −25°C until analysis.
Real-time and traditional RT–PCR
For rRT–PCR, a 1× master-mix (PrimeMix) containing real-time primers and probes specific for the universal detection of influenza A or an IPC RNA present within PrimeStore was utilized for detection according to the manufacturer's recommendations (Longhorn Vaccines & Diagnostics, USA). Amplification was performed using an ABI 7500 instrument (Life Technologies). RT–PCR thermocycling consisted of an RT incubation step at 50°C for 20 min followed by hot-start activation at 95°C for 5 min and 40 amplification cycles at 95°C for 15 s and 60°C for 32 s.
Traditional RT–PCR was performed using the SuperScript III High Fidelity One-Step kit (cat. no. 12574-035, Invitrogen, USA) according to the manufacturer's recommendations. Two primer pairs (forward 1204: 5′-TGTAAAACGACGGCCAGTAAGATGAAYACRCARTTCACAG-3′ and reverse 1778: 5′-CAGGAAACAGCTATGACCGTGTCAGTAGAAACAAGGGTGTTT-3′ and forward 379: 5′-TGTAAAACGACGGCCAGTACRTGTTACCCAGGRGATTTC-3′ and reverse 1204: 5′-CAGGAAACAGCTATGACCTCTTTACCYACTRCTGTGAA-3′) described by the World Health Organization [10] for amplification and sequencing influenza A(H1N1) 2009 field strains were used to generate a fragments with base-pair (bp) lengths of exactly 574 and 825 bp, respectively, from the H1N1 influenza haemagglutinin gene. Amplification was performed using an Applied Biosystems 2720 instrument (Life Technologies). RT–PCR thermocycling consisted of an RT incubation step at 50°C for 30 min followed by hot-start activation at 95°C for 2 min and 40 amplification cycles at 95°C for 15 s, 52°C for 15 s, and 68°C for 1 min. A final incubation of 68°C for 5 min for extension was performed. Influenza haemagglutinin gene fragments were visualized by UV illumination using 1·2% pre-made agarose gels stained with ethidium bromide (Invitrogen).
RESULTS
PrimeStore MTM microbial and viral inactivation
Microbial inactivation
PrimeStore MTM was shown to rapidly inactivate microbes including fungi, Gram-positive/-negative bacteria and viruses. Certified USP 32-NF 27<51> Antimicrobial Effectiveness Testing was performed using the membrane filtration technique for the quantitation of bacteria and fungi. At the first test period (24 h), 100% of bacteria and fungi were killed compared to the positive controls (Table 1 a). For these microbes, PrimeStore MTM met the inactivation criteria as described in USP Category 1 products (injections, emulsions, optic products, sterile nasal products, and ophthalmic products made with aqueous bases or vehicles). Additionally Bacillus subtilis spores were challenged using the method described in USP 51 to further evaluate PrimeStore MTM inactivation of microbial populations. B. subtilis spores were reduced by 99% within 24 h of exposure (Table 1 a). In a time-kill study of MRSA inoculated into PrimeStore MTM, viable bacteria were not detected (100% killing) at the earliest study time (5 min post-inoculation) or at any of the later evaluation times.
Table 1. PrimeStore Molecular Transport Medium inactivates: (a) bacteria, fungi and (b) viruses
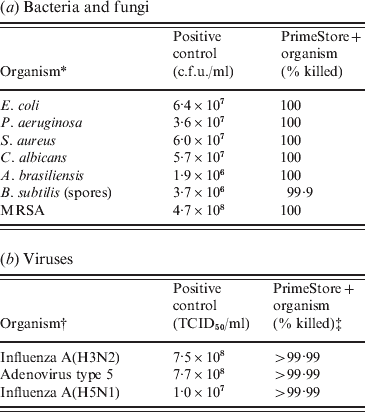
* All bacteria with the exception of MRSA were tested using the Membrane Filtration Technique. MRSA killing was performed using standard c.f.u. plate enumeration method.
† Influenza A(H3N2) inactivation testing performed by Virion Systems Inc. Highly pathogenic influenza A(H5N1) inactivation testing performed at Battelle Biomedical Research Center.
‡ Initial 4-log dilutions of PrimeStore MTM+viruses were required prior to inoculation into tissue culture lines due to PrimeStore MTM lysis of the MDCK tissue culture cells. Therefore, only a minimum 4-log TCID50/ml reduction was observable.
Viral inactivation
No viable virus influenza A(H3N2) or adenovirus type 5 was detected within seconds after being placed into PrimeStore MTM using viral loads as high as 107–108 TCID50. The recovery of highly pathogenic influenza A(A/Vietnam/1203/04) H5N1 virus at 108 TCID50/ml after incubation in PrimeStore MTM was equivalent to negative controls, indicating complete viral inactivation of H5N1 viruses (Table 1 b). Because PrimeStore MTM is designed to destroy membranes of cells and microbes, both MDCK and A549 cell lines were destroyed until the test samples were diluted 1:1000, LOD 103. Therefore, >4 log reductions (>99·99% killing) were documented in all spiked samples placed in PrimeStore MTM (Table 1 b).
PrimeStore MTM inactivates RNA/DNA nucleases
Nuclease digestion with RNase A, RNase T1, and DNase was used as a simple and effective method to examine PrimeStore protection of whole viral and single-stranded (ss)RNA. Influenza A virus inoculated in PrimeStore MTM was protected from exogenous addition of nucleases for 2 days at 37°C compared to commercial VTM, ethanol and other lysis buffers (Fig. 1). Assuming 100% PCR assay efficiency, rRT–PCR cycle threshold (CT) differences of 3·3 equate to an approximate tenfold change in initial template RNA, with a CT of 40 representing no detection [11]. RNA from influenza A virus (Fig. 1 a) and naked IPC ssRNA stored in PrimeStore MTM and incubated in a pool of nucleases (Fig. 1 b) exhibited a CT reduction of only 5·7 and 1·0, respectively, compared to almost complete digestion (CT values at or near 40) for other media after 48 h. This equates to preservation of about 104 viral copies compared to other media (Fig. 1).
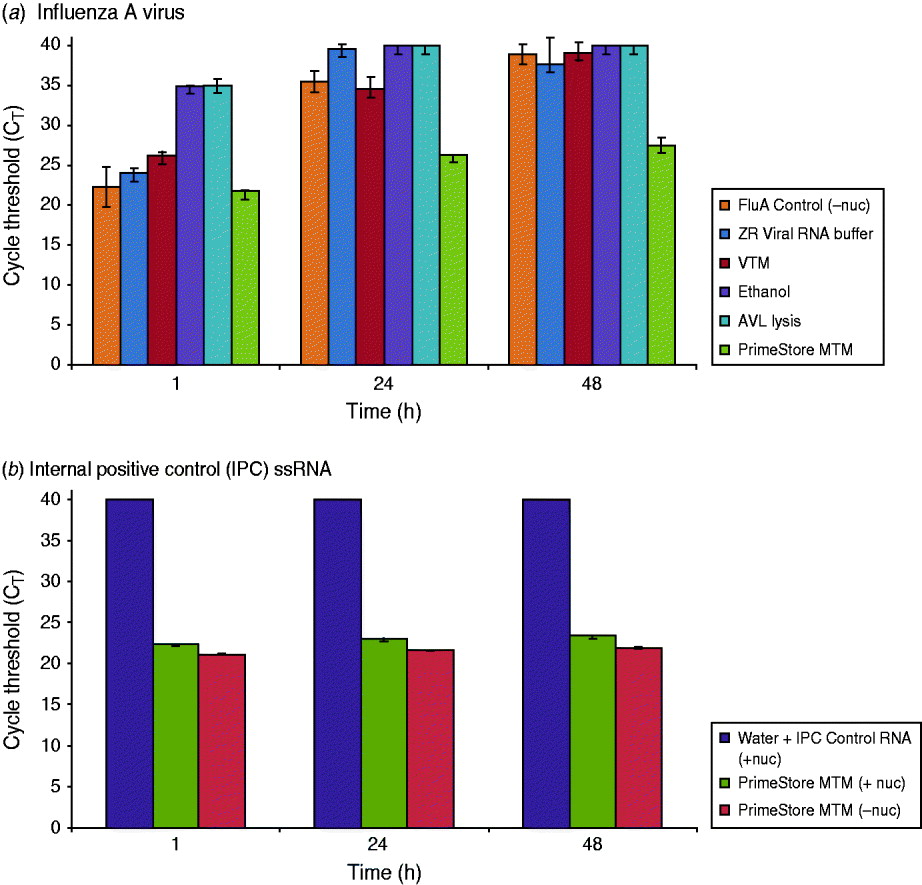
Fig. 1. PrimeStore MTM inactivates nucleases and preserves (a) influenza A viral RNA and (b) single-stranded internal positive control (IPC) RNA. Real-time RT–PCR analysis was performed using: 103 TCID50/ml whole influenza A(H3N2) virus preserved in PrimeStore MTM and other media* (a), and IPC RNA (b) after incubation in RNA/DNA nucleases (Ambion RNase A, T1, and Turbo DNase) at 37°C for 48 h. Nucleic acid extraction at 1, 24 and 48 h was performed and PCR amplified using PrimeMix Universal Influenza A and PrimeMix IPC assays (Longhorn Vaccines & Diagnostics). Mean and standard error bars for duplicate extractions are shown. A CT reading of 40 (CT=40) is no amplification. [* Viral transport media (VTM) is BD Universal Viral Transport Medium for Viruses, Chlamydiae, Mycoplasmas and Ureaplasmas (BD, USA); AVL Lysis Buffer is Buffer AVL viral lysis buffer (Qiagen, cat. no. 1014777; USA). ZR Viral RNA Buffer is from Zymo Research (cat. no. R1034-1-50).]
PrimeStore MTM enhances RNA stability and preservation
An influenza A(H1N1) reference strain (A/California/04/2009) was spiked into PrimeStore MTM containing a human clinical throat swab and incubated at 25°C (77 F) for 30 days. Influenza RNA from whole virus was highly preserved, with an average rRT–PCR CT reduction of 2·73 from day 0 baseline average at 25°C (Fig. 2).
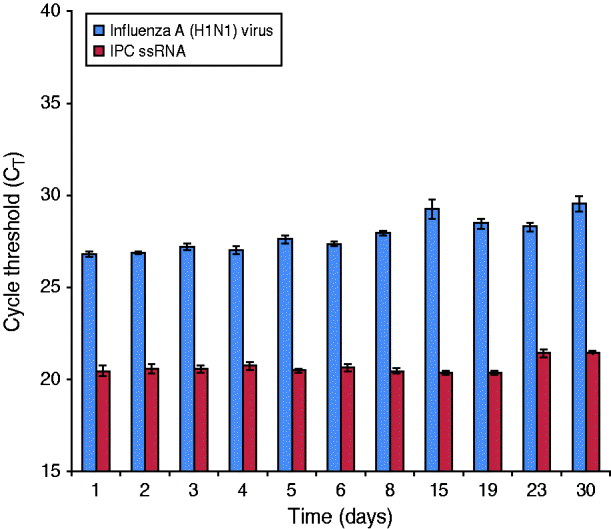
Fig. 2. PrimeStore stabilizes and preserves influenza A(H1N1) 2009 virus. A human clinical swab spiked with 5·3×102 TCID50/ml of influenza A(H1N1) A/California/04/2009 reference virus was placed into PrimeStore MTM and incubated at 25°C. Triplicate samples were extracted and analysed by real-time RT–PCR at the indicated time-point using an assay specific for influenza A virus and the internal positive control (IPC) single-stranded RNA piece present at fixed concentration (i.e. 0·02 pg) within PrimeStore MTM. Influenza H1N1-09 virus and single-stranded IPC RNA were preserved in PrimeStore MTM for 30 days with only minimal degradation (average 2·73 CT reduction) noted by day 30 for influenza virus at 25°C. Triplicate averages and standard error bars are shown.
Degradation of sample nucleic acids in PrimeStore MTM can be tracked through a non-specific, IPC ssRNA that is included in PrimeStore MTM at an initial concentration of 0·02 pg/μl. IPC RNA was preserved and stable at 25°C according to rRT–PCR amplification using a PrimeMix IPC rRT–PCR assay specific for the IPC RNA fragment (Fig. 2).
To demonstrate the preservation and stability of RNA polymers in PrimeStore MTM for downstream DNA sequencing, a stock strain (104 TCID50/ml) of A/California/04/2009 influenza A(H1N1) 2009 was spiked into a PrimeStore MTM and VTM and subsequently incubated at 38°C. Triplicate extractions at days 0, 2, 7, and 14 were performed for each time-point and subjected to standard and rRT–PCR amplification. Traditional RT–PCR was performed using primers to generate 574 bp and 825 bp products from the viral segment encoding the haemagglutinin protein (segment 4, ~1795 RNA bases) (Fig. 3 a). The 574 bp amplicon from influenza A H1N1 2009 virus preserved in PrimeStore MTM was visualized by agarose gel electrophoresis at all time-points compared to no amplification bands observed in extractions from influenza virus samples stored in standard VTM after day 0 (Fig. 3 a). The PCR band corresponding to the 825 bp amplification was evident to day 7 in virus stored in PrimeStore MTM™, compared to no amplification in virus stored in VTM after day 0 (Fig. 3 a).
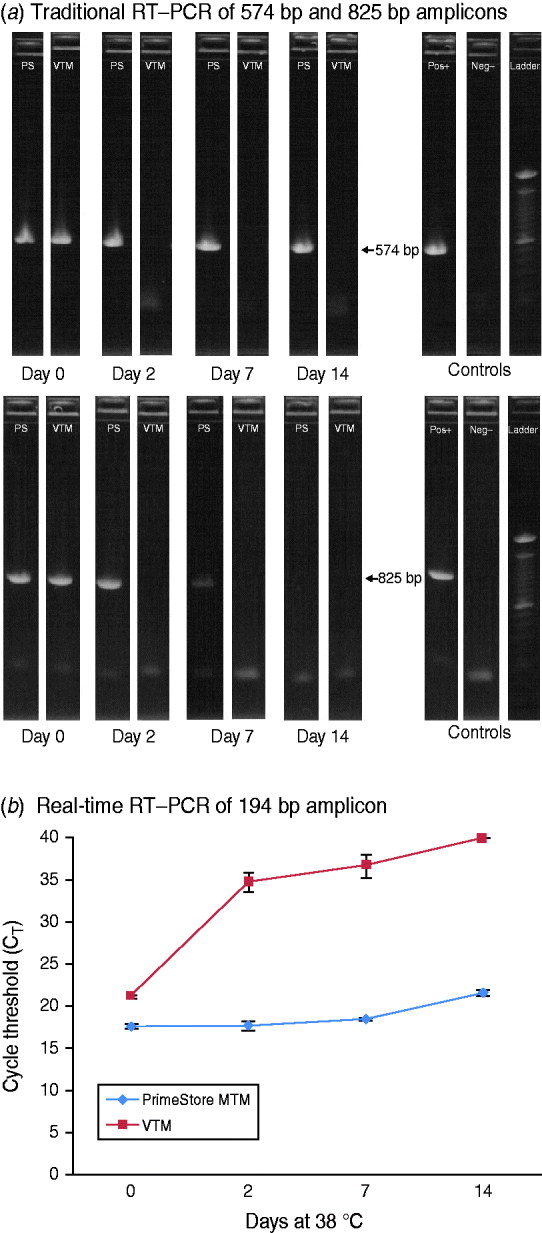
Fig. 3. RT–PCR amplification of influenza A(H1N1) 2009 stock virus in PrimeStore MTM and commercial VTM at 38°C using: (a) standard RT–PCR with primers to amplify 574-bp and 825-bp fragments and (b) real-time RT–PCR with primers to amplify a 194-bp fragment. A stock strain (104 TCID50/ml) of A/California/04/2009 was placed into PrimeStore MTM and VTM and incubated at 38°C for 0, 2, 7 and 14 days. Traditional RT–PCR amplification of 574 bp and 835 bp amplicons (using gel electrophoresis and UV visualization) were observed in PrimeStore preserved samples at days 14 and 7, respectively, compared to no amplification in VTM-preserved samples after day 0 (a). PrimeStore-preserved virus samples exhibited considerably less RNA degradation (average CT reduction=4·0) compared to no detection in VTM-preserved samples at day 14 (b). Reactions are average of triplicate with standard error shown. Commercial VTM is Copan UTM medium for Viruses, Chylaymida, Mycoplasma & Ureaplasma (Copan Diagnostics, Italy).
Similar results were observed using rRT–PCR for amplification of a 194-bp fragment specific for a conserved region of influenza A Matrix gene. Influenza virus preserved in PrimeStore showed considerably less degradation according to rRT–PCR (average CT reduction over 14 days=4·0) compared to no detection observed in virus stored in VTM (Fig. 3 b).
PrimeStore is compatible with commercial silica- and bead-based extraction kits
Commercial extraction kits are available in a variety of formats with most exploiting the basic principle of nucleic acid affinity for glass silicon dioxide particles present on filters or beads. To demonstrate PrimeStore MTM compatibility with commercial kits, reference influenza A(H1N1) virus (A/California/04/2009) was inoculated into PrimeStore containing a human throat swab specimen at two concentrations, 103 TCID50/ml and 101 TCID50/ml, and extracted using two commercial spin-column and two bead-based kits (Fig. 4). Viral influenza RNA in PrimeStore MTM was detected by rRT–PCR after extraction in all four commercial kits at high (103 TCID50/ml) and low (101 TCID50/ml) viral loads. Additionally, the IPC RNA (0·02 pg/μl) present in PrimeStore MTM was also detected by rRT–PCR after extraction using all four kits at levels consistent with the manufacturer's recommendations [12].
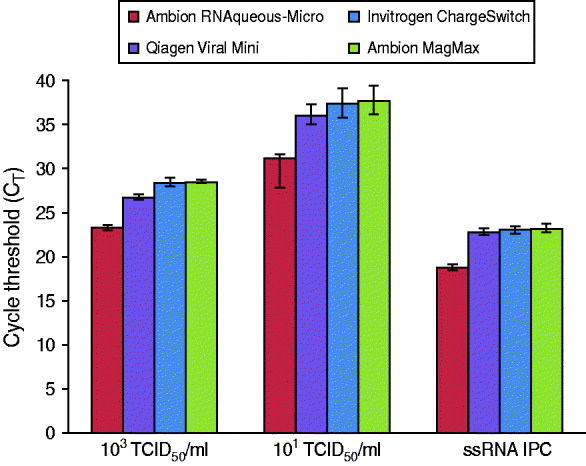
Fig. 4. PrimeStore molecular transport solution is compatible with commercial bench-top nucleic acid extraction kits including silica- and bead-based varieties. Two concentrations (103 TCID50/ml and 101 TCID50/ml), representing clinically relevant viral loads, i.e. a high and low, of stock H1N1 virus (A/California/04/2009) were inoculated into PrimeStore containing a clinical throat swab. Extractions were performed using two common silica- and bead-based nucleic acid kits (according to manufacturer's recommendations) and subjected to real-time RT–PCR analysis using an ABI 7500. Mean and standard error for triplicate extractions are shown.
DISCUSSION
Several companies such as Becton Dickinson, and Copan offer viral collection media for preserving viable organism for culture analysis. Unknown specimens collected in these media remain viable and are typically transported frozen to the laboratory and treated as biohazardous and potentially infectious. Therefore, there is considerable expense and risk of infection associated with the collection and transport of clinical specimens to reference laboratories.
Krafft et al. [Reference Krafft13] evaluated the use of respiratory samples fixed in ethanol as an alternative to commercial transport media using rRT–PCR. While ethanol may provide a suitable medium for influenza and other common upper respiratory pathogens, its use presents several other concerns including alcohol-shipping restrictions in many countries, high flammability, and the potential for incomplete lyses of certain microbes.
In a study by Blow et al. [Reference Blow14], viral RNA from field-collected arboviruses was stabilized at 32°C for 48 h using the AVL lysis buffer from a commercially available Qiagen extraction kit. While the samples are lysed and presumably non-infectious, the short, 2-day window of RNA preservation in Qiagen AVL makes this method unrealistic, even for routine pathogen surveillance.
The use of RNALater™ (Ambion, USA) has been reported in the literature as a solution for the preservation of collected specimens for molecular analysis [Reference Mutter15, Reference Florell16]. RNALater is a commercially available product designed for the preservation and maintenance of whole tissues for histology and microscopy of cells and microbes [17], but its use has also included the collection of field specimens for molecular analysis [Reference Nsubuga18–Reference McClure20]. RNALater contains a high concentration of ammonium sulfate for maintaining phospholipid integrity and intact cells. Several reports indicate that tissues and viruses from samples preserved in RNALater remain viable for extended periods of time and at a variety of temperatures [Reference Kurth21, Reference Uhlenhaut and Kracht22]. Maintaining viability of viruses and other microbes can be dangerous for many groups interested in safe, non-infectious transport of collected specimens for molecular-based diagnostics.
PrimeStore MTM was developed specifically to overcome these limitations in clinically collected specimens earmarked for routine downstream molecular diagnostics. Since PrimeStore MTM inactivates and kills a wide range of microbes (Table 1) it is well-suited for field collection in remote areas, triage centres, border crossings and during pandemics where cold-chain, transport and dissemination of potentially infectious pathogens are a concern. PrimeStore MTM lyses and inactivates microbial pathogens so they are non-viable; thus an alternative transport media is required for traditional culture. Preliminary data from a study in progress has demonstrated that PrimeStore MTM rapidly kills M. tuberculosis from clinical sputum samples (Dr Nazir Ahmed Ismail, Medical Microbiologist, University of Pretoria and NHLS, Pretoria, South Africa, personal communication). PrimeStore MTM is an ambient thermostable, proprietary molecular transport medium that rapidly inactivates nucleases (Fig. 1) and stabilizes and preserves released nucleic acids including labile RNA according to rRT–PCR analysis (Fig. 2). Furthermore, PrimeStore MTM stabilizes and preserves RNA polymers from hydrolysis and oxidation, and promotes thermostability at temperatures as high as 37°C for at least 14 days (Fig. 3).
A unique IPC ssRNA that is pre-mixed (3×105 target copies/μl) into PrimeStore MTM (and is thus stabilized) can be amplified by rRT–PCR to verify sample stability from the time of sample collection through extraction and detection (Fig. 2). The IPC RNA serves as a carrier species for low-level samples, controls for nucleic acid extraction and monitors sample integrity from the point of collection to detection. The RNA IPC in PrimeStore MTM is a synthetically produced (in vitro) 114-bp ssRNA polymer that is non-homologous by BLAST analysis to common upper respiratory pathogens and normal flora. A developed rRT–PCR assay (PrimeMix IPC, Longhorn Vaccines & Diagnostics) that targets this synthetic IPC RNA polymer is used as a uniplex rRT–PCR assay. A clinical collection medium containing exogenous IPC RNA fragments is important for ensuring integrity of transported or stored specimens for a wide variety of molecular-based detection systems.
PrimeStore MTM will also facilitate standard sequencing and meta-genomic analysis of original clinical samples by improving the quality of microbial nucleic acids in original specimens when they finally arrive in the laboratory. Recovery of RT–PCR amplification fragments over 1400 bases (data not shown) has been observed from viral RNA preserved and shipped in PrimeStore MTM at ambient temperature for several weeks. In harsh conditions, i.e. 38°C incubation, RT–PCR amplification of 574-bp and 825-bp fragments were observed from PrimeStore preserved virus where no amplification was observed from stock virus in commercial VTM (Fig. 3 a). The primers used to generate the 825-bp fragment represent the largest haemagglutinin gene fragment described by the WHO for influenza A(H1N1) sequencing [10]. Ambient temperature collection and preservation of large RNA fragments from clinical samples in PrimeStore MTM may be highly useful to groups who wish to perform direct DNA sequencing, genotyping, meta-genomic analysis and other NAT directly from original specimens.
In a 2007–2008 prospective paediatric clinical study, nasal washings preserved in PrimeStore MTM were subsequently analysed for influenza by rRT–PCR [Reference Daum23] and DNA sequencing of viral surface genes (Daum et al., unpublished observations). During the 2009 swine pandemic clinical throat swabs preserved in PrimeStore MTM were transported at ambient temperature and subjected to entire genome sequencing of swine influenza A(H1N1) virus using a Roche 454 system (Haung et al., unpublished data). In this work Haung et al. characterized four complete influenza genomes from the Dominican Republic, Columbia, Nicaragua, and Russia from swabs in PrimeStore MTM [GenBank accession numbers CY0Y3099-CY0Y3106, CY049833-CY049839, CY044152-CY044159, CY049896-CY049903; Haung et al., unpublished data].
PrimeStore MTM is compatible with many commercial extraction kits (Fig. 4). Nucleic acids are extracted directly from PrimeStore MTM according to standard manufacturer's protocol with only minor differences noted in CT values between column- and bead-based kits. Previous studies have shown that spin-column kits are slightly more robust compared to bead-based kits with certain sample types [Reference Cler24, Reference Dauphin25], but bead-based kits may be better when certain sample matrices, i.e. faecal and environmental are analysed. Additional studies are currently underway to evaluate the utility of PrimeStore MTM for use with other samples matrices such as faeces, blood, environmental samples and veterinary specimens. A 2010 influenza study that compared swabs in liquid Stuart's transport media (RTLS) and PrimeStore MTM from pigs experimentally infected with H1N1 influenza A virus found that virus from swabs in PrimeStore MTM and not Stuart's medium was detected at 5 days post-infection, a time when shedding is typically decreased or non-detectable [Reference Gramer26].
While PrimeStore MTM effectively lyses microbal pathogens, it is more than simply a lysis buffer as evident by comparison to typical lysis buffers from commercial extraction kits (Fig. 1). PrimeStore MTM is a blend of eight reagents that have been optimized to enhance RNA/DNA preservation after cellular lysis. PrimeStore MTM does not exhibit inhibition during nucleic acid extraction and PCR testing and has been shown to improve optimal recovery of influenza RNA (Fig. 3).
Notable reductions (4–5 CTs) in initial rRT–PCR CT values are routinely observed from virus inoculated in commercial VTM compared to equal amounts of virus in PrimeStore MTM (Fig. 3 b). This suggests that extraction and/or PCR inhibitors are present in commercial VTM that effect RNA detection during rRT–PCR. This may be problematical for groups who routinely extract RNA from original clinical samples collected in commercial VTMs for rRT–PCR or gene sequencing analysis. Accordingly, there is a need for an effective clinical VTM that serves the dual role of ensuring the viability of collected microorganisms for subsequent culture and propagation procedures and does not substantially interfere with downstream NAT processes.
In February 2010, PrimeStore MTM received FDA-Emergency Use Authorization as part of the complete Longhorn Influenza A/H1N1-09 Prime RRT–PCR Assay™ [12]. PrimeStore is the first molecular transport medium to receive EUA FDA approval, and the first to contain an IPC to control for specimen degradation from collection to detection.
ACKNOWLEDGEMENTS
We acknowledge the countless hours of effort from scientists, physicians and students alike who contributed to the development and clinical utility of PrimeStore MTM in molecular diagnostics. Specifically, Deena E. Sutter, M.D., Marty Ottolini, M.D., Aryeneesh K. Dotiwala, John Rodriquez, Demetra Kelenis, Jeff Fischer and Lou Schriefer.
DECLARATION OF INTEREST
Drs Luke T. Daum and Gerald W. Fischer and Ms. Susan A. Worthy are employees of Longhorn Vaccines & Diagnostics.









