Introduction
Placentation starts with the implantation of the human embryo into the maternal tissue and thus represents the key for successful pregnancy. Failures in placentation go along with complications in pregnancy such as preeclampsia, intrauterine growth restriction, placenta creta, and gestational trophoblastic disease often resulting in maternal or fetal death (Silva & Serakides, Reference Silva and Serakides2016). Methods for the histopathological analysis of implantation abnormalities in clinical routine are limited to lymphocyte infiltration, qualitative analysis of CD163, and hemosiderin staining (Ohyama et al., Reference Ohyama, Itani, Yamanaka, Goto, Kato, Ijiri and Tanaka2004; Roescher et al., Reference Roescher, Timmer, Erwich and Bos2014; Liu et al., Reference Liu, Ge, Ma, Zhang, Zhao and Cui2019). Research on implantation abnormalities is reduced to investigations of first to third trimester placentas, as pregnant woman are justifiably excluded for ethical reasons. However, immunostaining of placenta sections leads to new insights into fetal development and maternal pathophysiological pathways. In this context, immunostaining not only permits the evaluation of the histological architecture by preserving the tissue structure, it also enables the assessment of specific protein expression in the context of its microenvironment. However, reliable immunostaining is highly dependent on tissue preparation (Kim et al., Reference Kim, Roh and Park2016).
Whole-slide scanning technologies and automated quantitative image analyses have become standard methods used for histopathological diagnosis in clinics. Importantly, the artificial intelligence of the used programs requires constant training to continuously improve the recognition of specific pathologies (Gurcan et al., Reference Gurcan, Boucheron, Can, Madabhushi, Rajpoot and Yener2009; Gertych et al., Reference Gertych, Ing, Ma, Fuchs, Salman, Mohanty, Bhele, Velásquez-Vacca, Amin and Knudsen2015; Riordan et al., Reference Riordan, Varma, West and Brown2015). Hence, antibody staining needs to be consistently and precisely established, and research questions must not deviate from the target recognition of the analysis pipeline (Riordan et al., Reference Riordan, Varma, West and Brown2015). In basic research, however, research questions usually differ; hence, existing evaluation pipelines of automated quantitative image analysis programs often lack use. For this reason, conventional histological examinations in basic research, such as investigations of pregnancy-associated diseases and placenta pathologies, are still mainly limited to qualitative analyses, which are subjectively evaluated by the individual (Honig et al., Reference Honig, Rieger, Kapp, Dietl and Kämmerer2005; Gurcan et al., Reference Gurcan, Boucheron, Can, Madabhushi, Rajpoot and Yener2009; Higgins, Reference Higgins2015).
Thus, the present study provides a quantitative image analysis pipeline for an unbiased evaluation of fluorescent signal intensities of different antigen retrieval methods for 17 placenta-relevant antibodies in first trimester placental tissue. We additionally performed conventional qualitative immunostaining analysis to compare the differences in the results of the automated quantitative and qualitative evaluation methods.
Recently, placental organoid cultures and scaffold-based placental barrier and invasion models were developed to study placenta function and pathologies (Aengenheister et al., Reference Aengenheister, Keevend, Muoth, Schönenberger, Diener, Wick and Buerki-Thurnherr2018; Haider et al., Reference Haider, Meinhardt, Saleh, Kunihs, Gamperl, Kaindl, Ellinger, Burkard, Fiala, Pollheimer, Mendjan, Latos and Knöfler2018; Pemathilaka et al., Reference Pemathilaka, Reynolds and Hashemi2019). Many of these novel placental cell culture models require low-temperature embedding and antigen retrieval methods prior to immunostaining to preserve detailed structures of thermo-sensitive materials, such as biological matrices (Kreuder et al., Reference Kreuder, Bolaños-Rosales, Palmer, Thomas, Geiger, Lam, Amler, Markert, Lauster and Kloke2020), membranes of poly(ε-caprolactone)/poly(l-lactide) (PCL/PLA) (Vonbrunn et al., Reference Vonbrunn, Mueller, Pichlsberger, Sundl, Helmer, Wallner, Rinner, Tuca, Kamolz, Brislinger, Glasmacher and Lang-Olip2020), or mRNA (Fuchs et al., Reference Fuchs, Mueller, Daxböck, Stückler, Lang, Leitinger, Bock, El-Heliebi, Moser, Glasmacher and Brislinger2018). We have already investigated proteolytic pepsin digestion beneficial as an alternative to heat-induced antigen retrieval for thermo-sensitive scaffolds (Fuchs et al., Reference Fuchs, Mueller, Daxböck, Stückler, Lang, Leitinger, Bock, El-Heliebi, Moser, Glasmacher and Brislinger2018). Hence, the present study also enables us to investigate whether placenta-relevant antibodies positively tested after pepsin digestion on first trimester placenta tissue are applicable to scaffold-based 3D placenta cell culture samples. Based on this, we are able to histologically compare placenta-specific in vitro cell culture models to the in vivo situation.
Antibodies in the present work were chosen, as they are widely used in characterizing histological structures within placental tissue, both in research and in the diagnosis of placenta pathologies. These include markers detecting general structures as the cytoskeleton (β-actin) (Lanoix et al., Reference Lanoix, St-Pierre, Lacasse, Viau, Lafond and Vaillancourt2012), proliferating cells (pC) (Ki-67) (Gauster et al., Reference Gauster, Siwetz and Huppertz2009), vessels [CD31, CD34II, and von Willebrand factor (vWF)] (Kacemi et al., Reference Kacemi, Vervelle, Uzan and Challier1999), myofibroblasts in the villous stroma (desmin, vimentin, and SMA) (Kacemi et al., Reference Kacemi, Vervelle, Uzan and Challier1999; Sati et al., Reference Sati, Seval, Yasemin Demir, Kosanke, Kohnen and Demir2007), as well as markers for structures specific to placental villi such as cytotrophoblasts (CTs) [cytokeratin 7 (CK7), E-Cad, zonula occludens-1 (ZO-1), and alkaline phosphatase placental-like 2 (ALPPL2)] (Marzioni et al., Reference Marzioni, Banita, Felici, Paradinas, Newlands, Nictolis, Mühlhauser and Castellucci2001; Nuovo, Reference Nuovo2006; Gauster et al., Reference Gauster, Siwetz and Huppertz2009), syncytiotrophoblasts (SCTs) [beta-human chorionic gonadotropin(β-hCG) and leptin] (Londero et al., Reference Londero, Orsaria, Grassi, Calcagno, Marzinotto, Ceraudo, Fruscalzo, Driul and Mariuzzi2013; Tsai et al., Reference Tsai, Davis and Bryant-Greenwood2015), extravillous trophoblasts (EVTs) [human leukocyte antigen G (HLA-G)] (Guettler et al., Reference Guettler, Forstner, Cvirn, Maninger, Brugger, Nonn, Kupper, Pritz, Wernitznig, Dohr, Hutter, Juch, Isermann, Kohli and Gauster2021), and specific immune cells occurring in the placenta (CD163) (Böckle et al., Reference Böckle, Sölder, Kind, Romani and Sepp2008).
Hence, this study presents a comprehensive quantitative and qualitative analysis of placenta-relevant antibodies. It is intended to be a helpful tool for the selection of pretreatment and antibodies to improve research on placentation and implantation, such as investigations of pregnancy-associated diseases with first and third trimester placental tissues and the analysis of in vitro placenta models.
Materials and Methods
Sample Collection
Human first trimester placentas (gestational age: 9–10 weeks) were obtained from women undergoing elective pregnancy termination at the Department of Obstetrics and Gynaecology (Medical University of Graz) or the Femina Med Center (Graz) in accordance with the World Medical Association Declaration of Helsinki. All patients provided written informed consent, and ethical approval was granted by the Medical University of Graz (Nos. 31-019 ex 18/19 and 31-333 ex 18/19). The placental villi used for tissue-specific antibody detection were fixed in 3.7% paraformaldehyde for 24 h and paraffin-embedded according to standard histological procedures.
Low-Melting Paraffin Embedding of Thermo-Sensitive Electrospun PCL/PLA Scaffolds
Thermo-sensitive electrospun PCL/PLA scaffolds were cultured with first trimester placental villi or the human first trimester trophoblast cell line ACH-3P (Hiden et al., Reference Hiden, Wadsack, Prutsch, Gauster, Weiss, Frank, Schmitz, Fast-Hirsch, Hengstschläger, Pötgens, Rüben, Knöfler, Haslinger, Huppertz, Bilban, Kaufmann and Desoye2007) (a detailed description is provided in Supplementary Materials and Methods). Tissue cultured and cell-seeded scaffolds were fixed in 3.7% paraformaldehyde for 24 h (tissue cultured) or 15 min (cell-seeded) and then dehydrated and embedded by a recently published low-melting paraffin embedding method (Fuchs et al., Reference Fuchs, Mueller, Daxböck, Stückler, Lang, Leitinger, Bock, El-Heliebi, Moser, Glasmacher and Brislinger2018). Briefly, dehydration and low-melting-point paraffin embedding of fixed samples were performed with the KOS Microwave Multifunctional Tissue Processor (Milestone). Low-melting-point paraffin-embedded tissue-cultured and cell-seeded scaffolds were cut to 7 μm sections, transferred to SuperFrost Plus™ slides (Thermo Fisher Scientific), and dried overnight at 45°C.
Antigen Retrieval
To standardize immunofluorescent staining of each placental antibody, different antigen retrieval methods were performed on serial sections of first trimester placental villi: (a) no pretreatment, (b) pretreatment with pepsin (Sigma-Aldrich) (Fuchs et al., Reference Fuchs, Mueller, Daxböck, Stückler, Lang, Leitinger, Bock, El-Heliebi, Moser, Glasmacher and Brislinger2018), (c) heat-induced pretreatment with pH 6 citrate buffer, or (d) heat-induced pretreatment with pH 9 antigen retrieval solution (Leica Biosystems).
Sections included formaldehyde-fixed paraffin-embedded (FFPE) serial sections of first trimester placental villi (5 μm) and low-melting-paraffin serial sections of PCL/PLA scaffolds cultured with ACH-3P cells or first trimester placental villi. Sections were deparaffinized using Histolab Clear® solution (Histolab®) four times for 5 min each and rehydrated in a graded series of ethanol: 100, 96, 70, and then 50% ethanol, followed by three washes in distilled water for 3 min each.
For enzymatic antigen retrieval with pepsin, slides were incubated with 2 mg/mL pepsin (Sigma-Aldrich) in 0.1M HCl for 30 min at 37°C in a humidified chamber. The digestions were stopped by washing the slides with distilled water for 2 min. Heat-induced antigen retrieval was performed in a Decloaking Chamber (Biocare Medical). The deparaffinized placental villi FFPE sections were completely immersed in antigen retrieval solution or citrate buffer and steamed for 7 min at 120°C. Treated slides were cooled at room temperature for 20 min.
Immunofluorescence of First Trimester Placenta Sections
First trimester placenta sections were washed with 1× PBS and blocked with UV Block (Thermo Fisher Scientific) for 10 min. The primary antibody was diluted based on the antigen retrieval pretreatment and datasheet recommendations in antibody diluent (Agilent DAKO) and incubated for 30 min at room temperature. Antibodies used and their dilutions are listed in Table 1. Rabbit (X0936, 15 mg/mL, Agilent Technologies) and mouse (X0931, 0.1 mg/mL, Agilent Technologies) immunoglobulin fractions diluted in antibody diluent according to the highest antibody concentration of the corresponding species used in the study (X0936—noAR: 1:750, pepsin: 1:375, pH6:1:750, pH9:1:750; X0931—noAR: 1:20, pepsin: 1:10, pH6:1:20, pH9:1:20) served as negative controls after the different antigen retrieval methods. Next, slides were washed with 1× PBS and incubated with secondary antibody: Alexa Fluor 633-conjugated goat-anti-rabbit or goat-anti-mouse (1:200, Thermo Fisher Scientific, Catalog Nos. A21070 and A21050, detected in the Cy5 channel) for 30 min at room temperature. Finally, slides were washed, and nuclei were counterstained with DAPI for 5 min (1:2,000, Thermo Fisher Scientific). Sections were mounted with ProLong™ Gold Antifade Reagent (Thermo Fisher Scientific). The first section of each series was stained with hematoxylin and eosin to provide a histological overview (Fig. 2, columns a1–h1 and Fig. 3, columns a1–i1).
Table 1. List of Antibodies Used.

Origin, clone number, and stock concentrations are provided. Primary antibody concentrations used for each antigen retrieval pretreatment of first trimester placental tissue are gven.
* Headquarters locations of the manufacturers are listed in the Materials and Methods section of Supplementary material.
AR, antigen retrieval.
Immunofluorescence staining was additionally performed on untreated snap-frozen first trimester placenta cryosections to exclude the possibility of nonspecific binding on epitopes occurring through heat or enzymatic digestion and to verify antibody specificity. More detailed information on snap freezing of first trimester placenta tissue, sectioning, and immunostaining is available in the Supplementary Materials and Methods.
Immunofluorescence of Cell-Seeded and Tissue-Cultured PCL/PLA Scaffold Sections
Antibodies showing positive staining on first trimester placental sections after enzymatic antigen retrieval with pepsin (summarized in Supplementary Table 1) were tested on PCL/PLA scaffolds cultured with ACH-3P cells or first trimester placental tissue. Only antibody-relevant proteins that were likely to be expressed by ACH-3P cells (Hiden et al., Reference Hiden, Wadsack, Prutsch, Gauster, Weiss, Frank, Schmitz, Fast-Hirsch, Hengstschläger, Pötgens, Rüben, Knöfler, Haslinger, Huppertz, Bilban, Kaufmann and Desoye2007) were tested for their antibody binding specificity on PCL/PLA scaffolds (marked in Supplementary Table 1). Cell-seeded and tissue cultured PCL/PLA samples underwent pepsin digestion for antigen retrieval and immunofluorescent staining as described above. Rabbit (X0936, 15 mg/mL, Agilent Technologies) and mouse immunoglobulin fractions (X0931, 0.1 mg/m, Agilent Technologies) diluted in antibody diluent according to the highest used antibody concentration of the corresponding species (X0936—pepsin: 1:1,500, X0931—pepsin: 1:10) served as negative controls.
Image Acquisition
Images were acquired by scanning 36 tiles at 20× magnification, corresponding to a total area of 6.4 mm2 (Fig. 1a), over the same spatial area of all serial sections of the tested antibody. Brightfield and fluorescence images were acquired using the Zeiss Observer.Z1 inverted microscope (Carl Zeiss, Oberkochen, Germany) equipped with the Colibri 7 LED illumination system (Carl Zeiss). For brightfield images, the Axiocam 506 camera (Carl Zeiss) was used, and fluorescence images were taken with the Axiocam 702 mono (Carl Zeiss) equipped with an excitation and emission filter set for the visualization of DAPI, FITC, Cy3, and Cy5. The 20× objective (LD Plan-Neofluar 20×/0.40 corr., D = 0–1.5 mm; Carl Zeiss), and the ZEN 3 blue software (Version 3.0; Carl Zeiss) was used for capturing both the brightfield and fluorescence images.
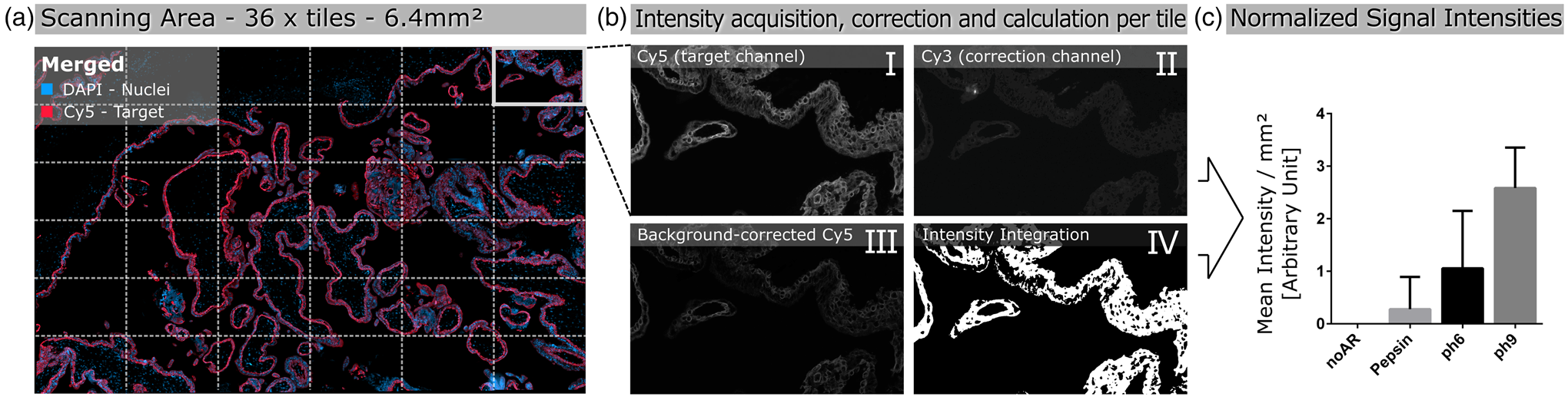
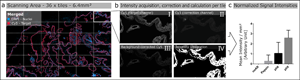
Fig. 1. Schematic illustration of automated quantitative fluorescent image analysis. (a) A 6.4 mm2 area resulting from 36 non-overlapping 20× images was captured. Images were taken in four fluorescent channels: DAPI, FITC, Cy3, and Cy5 (merged image only shows DAPI and Cy5). (b) Antibody staining fluoresced in the target channel Cy5 (I). Cy3 channel autofluorescence was used as correction (II). Background correction was done by subtracting Cy3 from the target channel Cy5 (III). Using Cy5 image masks (IV) across the background-corrected images, the mean signal intensity over the whole scanning area was calculated in arbitrary units with CellProfiler software. (c) Mean signal intensity was normalized over an area of 1 mm2 and plotted for the corresponding pretreatment (the graph displays an example). The analysis (a–c) was conducted for each pretreatment across all 17 antibodies.
Quantitative Image Analysis
The Cy5 channel (Fig. 1b-I) exposure time was set using the “Auto Exposure” function on the most intense staining of each antibody out of all different antigen retrieval methods, providing the shortest possible exposure time. The same exposure time was used for all antigen retrieval treatments for each antibody to enable comparison of the fluorescence intensities within a group. A constant exposure time was used for FITC and CY3 channels (Fig. 1b-II) to detect background fluorescence.
We used the open-source cell image analysis software CellProfiler (version 3.1.8; McQuin et al., Reference McQuin, Goodman, Chernyshev, Kamentsky, Cimini, Karhohs, Doan, Ding, Rafelski, Thirstrup, Wiegraebe, Singh, Becker, Caicedo and Carpenter2018) to evaluate the mean signal intensity of the Cy5 channel over the entire 6.4 mm2 region corresponding to ~2.7 billion pixels (Fig. 1a). Specific, positive antibody staining exclusively appears in the wavelength range that corresponds to the fluorophores conjugated to the secondary antibody (Weibrecht et al., Reference Weibrecht, Lundin, Kiflemariam, Mignardi, Grundberg, Larsson, Koos, Nilsson and Söderberg2013). Signals detected in multiple channels were classified as false-positive and, therefore, nonspecific staining. To exclude false-positive signals from the analysis, the Cy3 channel was subtracted from the Cy5 channel (Fig. 1b-III). Additionally, existing background noise was eliminated by using a threshold to improve the signal-to-background ratio (Fig. 1b-IV). Mean signal intensities were normalized across a 1 mm2 area and plotted for the respective pretreatments for each antibody (Fig. 1c). More detailed information on the custom-designed CellProfiler pipeline is available in Supplementary Materials and Methods. Calculations and detailed values of the quantitative image analysis are given in Supplementary Table 2.
Morphological Analysis
In addition to the automated quantitative image analysis, immunofluorescent staining of first trimester placenta was qualitatively evaluated. The histological classification of various cell types occurring in first trimester placentas was performed regarding the specific staining reaction pattern and the presence of nonspecific background for each antibody. Positive antibody staining was counted as specific when the fluorescence signal exclusively appeared in the wavelength range corresponding to the fluorophores conjugated to the secondary antibody. Signals appearing in multiple channels were classified as nonspecific staining. Individual cell types including CTs, SCTs, endothelial cells (ECs), EVTs, fibroblasts (Fib), smooth muscle cells, Hofbauer cells , and pC were evaluated. Additionally, a classification category for nonspecific staining was introduced, marked in red to classify irregular and insufficient antibody staining all over the tissue. Green coloring was used to mark specific staining pattern. An intensity grading system was applied, using “-” for negative staining, “○” to display a positive staining reaction, and “●” indicating the cell type that appears the brightest within an antigen retrieval group all over the tissue (if there are intensity differences between cell types). “◉” marks cell types that show a brighter signal in only some parts of the tissue compared to other positive stained cell types within an antigen retrieval treatment group. Results of the qualitative evaluation are shown in Figure 4.
Statistical Analyses
Mean intensities resulting from the automated quantitative image analysis were statistically evaluated using GraphPad PRISM software (version 6.01). Outliers were identified using the ROUT test with a Q value of 1% (Motulsky & Brown, Reference Motulsky and Brown2006). Cleaned data were used for further analysis. Data were tested for Gaussian distribution with a Kolmogorov–Smirnov test. Mean fluorescence intensities of the Cy5 signal of all antigen retrieval methods were compared using one-way ANOVA with Tukey's multiple comparison test. Results were considered statistically significant when p < 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
Results
Automated Quantitative Image Analysis Revealed Differences in Immunofluorescent Intensities Based on the Selected Antigen Retrieval Method
Automated quantitative image analysis revealed that 9 out of 17 tested antibodies required antigen retrieval for immunofluorescent staining (Figs. 2, 3). Antibodies specific for CK7 (rabbit), E-cadherin, vWF, ZO-1, CD31, HLA-G, Ki-67, leptin, and CD163 needed selected pretreatments to stain positive for their antigen. However, β-Actin, desmin, smooth muscle actin (SMA), vimentin, ALPPL2, β-hCG, CD34II, and CK7 (mouse) showed specific immunofluorescent signals without any antigen retrieval pretreatment.

Fig. 2. Immunostaining after different antigen retrieval pretreatments on first trimester placenta serial sections. Antibodies tested show specific binding after enzymatic digestion with pepsin. Hematoxylin and eosin staining (columns a1–h1) provides a morphological overview of the tissue. Each of the fluorescent-stained serial sections was treated using one of the different methods: none, pepsin digestion, pH 6 heat-induced epitope retrieval, or pH 9 heat-induced epitope retrieval. Columns 2–5 show representative image details of antibody-stained serial sections after the different pretreatments. The mean fluorescence intensity of the target per mm2 evaluated by quantitative automated image analysis is given in the bar chart for each pretreatment (columns a6–h6). β-hCG (b3, b6) and vWF (g3, g6) show significantly improved staining after pepsin digestion. Detailed values of the quantitative image analysis are given in Supplementary Table 2. Scale bars represent 50 μm. noAR, no antigen retrieval; pepsin, enzymatic digestion with pepsin; pH 6, heat-induced epitope retrieval with pH 6 citrate buffer; pH 9, heat-induced epitope retrieval with pH 9 antigen retrieval buffer.

Fig. 3. Immunostaining after different antigen retrieval pretreatments on first trimester placenta serial sections. Tested antibodies show no specific staining after enzymatic pepsin digestion. Hematoxylin and eosin staining (columns a1–i1) provides a morphological overview of the tissue. Each of the fluorescent-stained serial sections was treated using one of the different methods: none, pepsin digestion, pH 6 heat-induced epitope retrieval, or pH 9 heat-induced epitope retrieval. Columns 2–5 show representative image details of antibody-stained serial sections after the different pretreatments. The mean fluorescence intensity of the target per mm2 evaluated by quantitative automated image analysis is given in the bar chart for each pretreatment (columns a6–i6). Anti-CD163 was the only antibody (i3) displaying nonspecific staining over the entire tissue after pepsin digestion. Detailed values of the quantitative image analysis are given in Supplementary Table 2. Scale bars represent 50 μm. noAR, no antigen retrieval; pepsin, enzymatic digestion with pepsin; pH 6, heat-induced epitope retrieval with pH 6 citrate buffer; pH 9, heat-induced epitope retrieval with pH 9 antigen retrieval buffer.
We found heat induction to be the most efficient form of antigen retrieval, followed by enzymatic digestion. Heat-induced antigen retrieval was strongly influenced by the pH value. Four of the antibodies examined, CD34II (Figs. 2c4, 2c6), CD31 (Figs. 3b4, 3b6), leptin (Figs. 3f4, 3f6), and CD163 (Figs. 3i4, 3i6), displayed the most intense staining after heat-induced antigen retrieval with pH 6. Eleven placenta-specific antibodies, ALPPL2 (Figs. 2a5, 2a6), CK7 (rabbit; Figs. 2d5, 2d6), CK7 (mouse; Figs. 2e5, 2e6), E-cadherin (Figs. 2f5, 2f6), ZO-1 (Figs. 2h5, 2h6), β-Actin (Figs. 3a5, 3a6), desmin (Figs. 3c5, 3c6), HLA-G (Figs. 3d5, 3d6), Ki-67 (Figs. 3e5, 3e6), SMA (Figs. 3g5, 3g6), and vimentin (Figs. 3h5, 3h6), showed the most intense staining after pretreatment with pH 9.
Enzymatic antigen retrieval with pepsin yielded the brightest signals for β-hCG (Figs. 2b3, 2b6) and vWF (Figs. 2g3, 2g6). Overall, pretreatment for antigen retrieval was necessary to get the highest signal intensities of selected antibody within its group.
Negative controls (rabbit and mouse immunoglobulin fractions) showed no expression of the antibodies used. Images are shown in Supplementary Figures 2a1–2a4 and 2b1–2b4.
Antibody Binding Specificity Is Influenced by the Antigen Retrieval Method
We used first trimester placental tissue to investigate the influence of antigen retrieval on antibody binding specificity. Qualitative assessment clearly demonstrated that the method of antigen retrieval had implications for antibody binding specificity and signal intensities (Fig. 4). In some cases, e.g. CK7 antibodies, the determination of cell-type specificity was not possible, since intense staining was frequently observed in two cell types over large areas.

Fig. 4. Qualitative evaluation of antibody staining of different cell types in first trimester placenta after different antigen retrieval pretreatments. - indicates no staining, red indicates nonspecific staining, green indicates specific staining. ○ marks cell types that shows positive staining within an antigen retrieval group. ● marks the cell type that show the most intense staining compared to other positive cell types within an antigen retrieval group. ◉ marks cell types that show more intense staining in some parts of the tissue compared to other stained cell types within an antigen retrieval group. noAR, no antigen retrieval; CT, cytotrophoblast; SCT, syncytiotrophoblast; EC, endothelial cells; EVT, extravillous trophoblast; Fib, fibroblasts; SMC, smooth muscle cells; HBC, Hofbauer cells; pC, proliferating cells.
Pretreatment with Pepsin Increased Staining Intensities of β-hCG and vWF Antibodies
All examined antibodies displaying successful staining after enzymatic antigen retrieval with pepsin are illustrated in Figure 2. The quantitative image analysis of the immunofluorescent staining of β-hCG and vWF after pepsin digestion showed significantly higher mean fluorescent signal intensities compared to all other antigen retrieval methods (Figs. 2b6, 2g6), corresponding to the qualitative evaluation (Fig. 4).
CD163 Showed Nonspecific Staining after Enzymatic Antigen Retrieval
While the enzymatic digestion with pepsin improved the staining intensity and/or specificity of certain antibodies, other epitopes were completely degraded, preventing antibody binding. As shown in Figure 3 (columns a3–h3), eight antibodies completely lacked staining after pepsin pretreatment. Anti-CD163 was the only antibody (Fig. 3i3) displaying nonspecific staining over the entire tissue after pepsin digestion.
ACH-3P Cells Cultured on PCL/PLA Scaffolds Stained Positive after Enzymatic Antigen Retrieval with Pepsin
The ACH-3P cell line was generated by fusing first trimester trophoblasts to the choriocarcinoma cell line AC1-1. As such, it is characterized by the expression of proteins specific for EVTs (Hiden et al., Reference Hiden, Wadsack, Prutsch, Gauster, Weiss, Frank, Schmitz, Fast-Hirsch, Hengstschläger, Pötgens, Rüben, Knöfler, Haslinger, Huppertz, Bilban, Kaufmann and Desoye2007). Only proteins known to be expressed by trophoblast cells as β-Actin, CK7 (rabbit and mouse origins), E-cadherin, and ZO-1 were tested on these sections (see Supplementary Table 1).
Positive staining after enzymatic antigen retrieval with pepsin was obtained after staining for β-hCG, CK7 (rabbit and mouse origins), E-cadherin, and ZO-1 (Fig. 5). Qualitatively, β-hCG (Figs. 5a, 5b) and ZO-1 (Figs. 5i, 5j) showed moderate staining, while high staining intensities were observed for CK7 (rabbit; Figs. 5c, 5d), CK7 (mouse; Figs. 5e, 5f), and E-cadherin (Figs. 5g, 5h). The mouse-origin anti-CK7 antibody showed more consistent staining and improved morphological cell integrity on PCL/PLA scaffolds cultured with ACH-3P compared to the rabbit-origin anti-CK7 antibody. Rabbit and mouse immunoglobulin fractions showed no staining of the antibodies used.

Fig. 5. Antibody staining of the human first trimester trophoblast cell line ACH-3P cultured on heat-sensitive PCL/PLA with placenta-relevant antibodies after pepsin digestion. White boxes indicate the zoom-in area on the right image side. All antibodies tested showed specific fluorescence signals. Scale bars in (a,c,e,g,i,k,m,o) represent 100 μm, and those in (b,d,f,h,j,l,n,p) represent 20 μm.
Positive Staining of Endothelial and Trophoblast Cells of First Trimester Placental Villi Cultured on PCL/PLA Scaffolds Was Observed after Pepsin Digestion
To test the applicability of the enzyme-based immunostaining protocol for thermo-sensitive samples, first trimester placental villi cultured on PCL/PLA scaffolds underwent enzymatic antigen retrieval with pepsin and antibody staining. All antibodies tested on these samples showed positive immunostaining.
The anti-CD34II antibody displayed specific staining of EC, marking blood vessels in the villous stroma (Figs. 6a, 6b). Positive signals of the anti-ALPPL2 antibody were observed exclusively in SCT cells and migrated EVTs (Figs. 6c, 6d). Both rabbit- and mouse-origin anti-CK7 antibodies positively stained scaffold-attached first trimester placental villi. Qualitatively, the mouse-origin anti-CK7 antibody (Figs. 6g, 6h) showed brighter and more distinct fluorescence compared to the rabbit-origin anti-CK7 antibody (Figs. 6e, 6f), which displayed impaired morphological cell integrity. However, both antibodies positively recognized EVTs within the PCL/PLA scaffold.
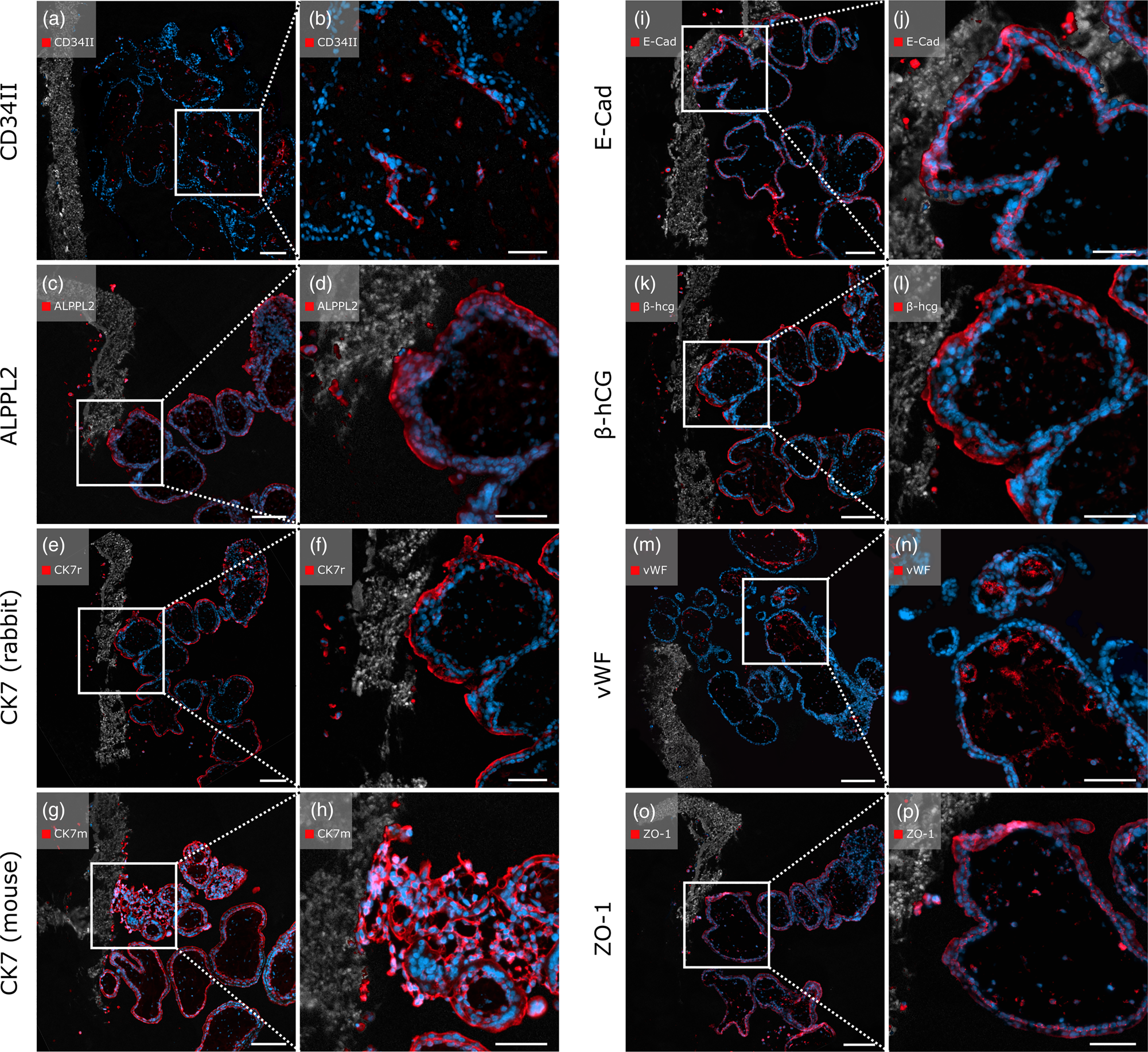
Fig. 6. Immunofluorescent staining with placenta-relevant antibodies on first trimester placental villi attached to heat-sensitive PCL/PLA scaffolds after pepsin digestion. White boxes indicate the zoom-in area on the right image side. All antibodies tested showed specific fluorescence signals. Scale bars in (a,c,e,g,i) represent 100 μm, and those in (b,d,f,h,j) represent 50 μm.
In addition, positive staining was observed for the transmembrane protein E-cadherin. Both the CT cells and the migrating EVTs showed positive fluorescence (Figs. 6i, 6j). As expected, SCTs were positive for β-hCG, as were migrated EVTs within the membrane (Figs. 6k, 6l). The endothelial marker vWF displayed positive staining specifically on EC marking blood vessels within the villous stroma. EVTs that migrated through the PCL/PLA scaffold appeared negative for vWF (Figs. 6m, 6n). Anti-ZO-1 antibody showed positive staining with specific fluorescent signals of several cell types, including ECs, CTs, SCTs, EVTs, and F (Figs. 6o, 6p). Rabbit and mouse immunoglobulin fractions showed no expression of the antibodies used (Supplementary Figs. 2c1, 2c2).
Pepsin Digest Serves as Heatless Antigen Retrieval Alternative for Placenta-Relevant Antibodies on Scaffold-Based Placental Cell Culture Samples
All antibodies positively tested after pepsin digestion on first trimester placenta tissue (Fig. 2, columns a3–h3) showed positive immunostaining on tissue cultured PCL/PLA samples after enzymatic antigen retrieval (Fig. 6). In addition, all antibodies expected to be expressed by the ACH-3P cell line displayed positive fluorescence signals on ACH-3P cultured PCL/PLA samples after enzymatic antigen retrieval (Fig. 5).
Immunofluorescent Staining of Snap-Frozen First Trimester Placenta Cryosections without Antigen Retrieval Verifies Antibody Specificity
To exclude nonspecific binding due to heat or enzymatic antigen retrieval, antibodies were additionally tested on first trimester placental cryosections without antigen retrieval. All tested antibodies showed positive immunostaining on the expected cell types and morphological structures in accordance with the references of the literature search (Supplementary Fig. 1; Supplementary Table 3). The comparison of the immunostaining on untreated cryosections with the staining on heat or enzymatically treated paraffin sections allowed the detection or exclusion of nonspecific binding after antigen retrieval and thus the verification of binding specificity. Comparing the results in a qualitative way, no unspecific antibody binding could be detected after immunostaining on antigen retrieval treated sections. Negative controls (rabbit and mouse immunoglobulin fractions) showed no expression of the antibodies used (Supplementary Figs. 1o, 1p).
Discussion
In this study, we present an evaluation of antigen retrieval methods for 17 antibodies widely used in placentation and implantation diagnostic and research. We established an automated quantitative image analysis method for the quantification of fluorescent signal intensities over large image areas and compared the results with conventional qualitative histological analysis methods. In contrast to qualitative histological examinations usually evaluated by an individual (Honig et al., Reference Honig, Rieger, Kapp, Dietl and Kämmerer2005), the major advantage of automated computational analysis is a more objective and therefore less biased interpretation of data (Nezami & Butcher, Reference Nezami, Butcher, Golstein and Hersen2000; Fantuzzo et al., Reference Fantuzzo, Mirabella, Hamod, Hart, Zahn and Pang2017). Our quantitative evaluation allows for a more precise description of immunostaining as it is based on large portions of the tissue compared to small sections, which are often used in conventional quantitative analyzed histological studies.
One purpose of antigen retrieval is to reveal epitopes and other cellular targets (Robinson & Vandré, Reference Robinson and Vandré2001). This is necessary because tissue samples are generally fixed prior to histological processing. Fixation preserves the biological material as close as possible to its natural state (Robinson & Vandré, Reference Robinson and Vandré2001), since primary and secondary protein structures are cross-linked (Mason & O'Leary, Reference Mason and O'Leary1991). However, sample fixation significantly reduces the immunoreactivity of most antigens (Battifora & Kopinski, Reference Battifora and Kopinski1986; Leong & Gilham, Reference Leong and Gilham1989).
With the successful implementation of a comprehensive, automated quantitative image analysis pipeline, we identified the optimal pretreatment for selected placenta-relevant antibodies. Our approach allowed for a direct comparison of the effects of different antigen retrieval pretreatments on immunostaining specificity and intensity. While each antibody showed specific fluorescence signals after heat-induced antigen retrieval; however, not all markers displayed positive staining after proteolytic digestion with pepsin, e.g. nonspecific staining was observed with CD163. We assume that this is due to incomplete degradation of cross-links, as nonspecific primary antibody binding may occur in the presence of unreacted hydroxymethyl groups after formaldehyde fixation. Additionally, depending on the type of epitope and marker, nonspecific interactions may occur between the primary antibody and other antigens within the tissue (Daneshtalab et al., Reference Daneshtalab, Doré and Smeda2010). No staining was observed with eight of the antibodies after pepsin digestion, indicating that their epitopes may have been completely degraded. This closely parallels findings by Battifora and Kopinski (Reference Battifora and Kopinski1986) and Daneshtalab et al. (Reference Daneshtalab, Doré and Smeda2010), who argued that overdigestion damages the morphology and immunoreactivity of tissue and may reduce the number of antigenic sites available for immunostaining. We speculate that the threshold for overdigestion varies and depends not only on the duration of fixation but also on the marker and the type of tissue examined. As such, this must be empirically determined for each antibody. It is possible that shortening the digestion time for antibodies not showing immunostaining signals using proteolytic antigen retrieval would yield positive results. These speculations are consistent with other research groups (Daneshtalab et al., Reference Daneshtalab, Doré and Smeda2010).
To exclude nonspecific staining due to enzymatic or heat-induced antigen retrieval, we performed antibody staining on first trimester placenta cryosections as an additional verification method. As cryosections enable immunostaining without antigen retrieval techniques, the occurrence or loss of nonspecific antigenicity is unlikely (Hira et al., Reference Hira, Jong, Ferro, Khurshed, Molenaar and van Noorden2019). This approach thus provides a valid method verifying specificity of the antibodies used in this publication, besides a comprehensive literature study to determine whether relevant morphological structures are expressed in first trimester placental tissue (MacNeil et al., Reference MacNeil, Vathiotis, Martinez-Morilla, Yaghoobi, Zugazagoitia, Liu and Rimm2020). However, the tissue morphology of snap-frozen placenta samples was of inferior morphological quality compared to paraffin-embedded samples due to the freezing procedure (Blaschitz et al., Reference Blaschitz, Gauster and Dohr2008). Thus, the requirements of tissue conservation through fixation and the preservation of antigenicity in immunostaining constantly conflict. Nevertheless, accurate identification of immunostaining on cryosections was possible. All antibodies showed positive staining in the expected morphological areas. The comparison of the immunostaining on untreated cryosections with the staining on heat- or enzyme-treated paraffin sections revealed that all antibodies bind specifically in the expected regions. In the case of CD163, comparison with staining on cryosections again demonstrated that immunostaining after pepsin digestion results in nonspecific binding.
To further ensure antibody specificity, monoclonal antibodies should be preferred, as they bind to a single, definable epitope on the target antigen and are highly specific and consistent between experiments (MacNeil et al., Reference MacNeil, Vathiotis, Martinez-Morilla, Yaghoobi, Zugazagoitia, Liu and Rimm2020). Thus, in the present study, 11 of the 17 antibodies investigated derived from monoclonal origins.
Our study was supplemented by a qualitative analysis which revealed that the cell type-specific staining for each antibody depends on the applied antigen retrieval method, as observed for β-hCG and both anti-CK7 antibodies. Based on that observation, it is important to relate signal intensities to cell type-specific staining efficiency. For example, high fluorescence intensity levels in quantitative analysis either can occur as high-intensity signal originating from one cell type, or are a result of accumulating signal intensities based on staining of multiple, different cell types. Thus, the qualitative aspect should always be considered before interpreting quantitative image analysis to avoid the misinterpretation of fluorescence signals. This highlights an important limitation of computer-based image quantification. While artificial intelligence and deep-learning systems have revolutionized global pathology in the primary diagnosis of certain cancer cell types, computational algorithms in other histological fields, especially in unsupervised architectures, are prone to misinterpreting artifacts as true biological signals. Important quality parameters that matter can be inconspicuous and easily overlooked (Evans et al., Reference Evans, Salama, Henricks and Pantanowitz2017; Tomaszewski, Reference Tomaszewski and Cohen2021).
Nevertheless, quantification in histology and thereby standardized quantitative automated image analysis are becoming increasingly important in the diagnosis of placenta-associated diseases. In addition to studies dealing with the semi-quantitative evaluation of syncytial knots to investigate placental malfunction (Senagore et al., Reference Senagore, Holzman, Parks and Catov2016; Kidron et al., Reference Kidron, Vainer, Fisher and Sharony2017; Parks, Reference Parks, Yee Khong, Mooney, Nikkels, Morgan and Gordijn2019), the quantification of the vessel number and layer composition during uterine and placental vascularization, for instance, is of great importance for diagnosing pathologies such as preeclampsia or intrauterine growth restriction. Up to now, the visualization of placental vascularization is mainly performed using 3D ultrasound technology (Campbell, Reference Campbell2007), which has limited precision in depicting the histological composition of vessel layers. As a result, important information at a cellular level remains unknown. To generate further knowledge regarding these pregnancy-associated diseases, automated quantitative image analysis based on immunostained placental sections using markers such as desmin, vimentin, and SMA for vascular detection, as described in this study, might provide valuable insights. Beyond that, the evaluated antibodies offer the possibility for further placenta-related histological investigations on a quantitative level, such as trophoblast invasion by using the trophoblast markers HLA-G, CK7, and β-hCG or the examination of maternal and childhood obesity via ALPPL2 and leptin antibodies (Hirschmugl et al., Reference Hirschmugl, Crozier, Matthews, Kitzinger, Klymiuk, Inskip, Harvey, Cooper, Sibley, Glazier, Wadsack, Godfrey, Desoye and Lewis2018).
Automated quantitative image analysis is a powerful technique minimizing operator error and biased interpretation; however, the qualitative assessment of antibody binding remains essential to properly assess immunofluorescence specificity and to avoid misinterpreting the data (Nezami & Butcher, Reference Nezami, Butcher, Golstein and Hersen2000; Gertych et al., Reference Gertych, Ing, Ma, Fuchs, Salman, Mohanty, Bhele, Velásquez-Vacca, Amin and Knudsen2015). Hence, in our opinion, the qualitative analysis of each antibody enables the optimization of quantitative analysis methods and thus the possibility of objective diagnostic procedures.
In addition, our automated quantitative image analysis approach enabled a direct assessment of whether pepsin digestion can serve as an alternative to conventional antigen retrieval pretreatment methods for thermo-sensitive scaffold-based placental cell culture systems. Proteolytic digestion with pepsin expands on a recently described method enabling antigen retrieval of thermo-sensitive specimens as an alternative to common heat-induced methods (Ramos-Vara and Beissenherz, Reference Ramos-Vara and Beissenherz2000; Fuchs et al., Reference Fuchs, Mueller, Daxböck, Stückler, Lang, Leitinger, Bock, El-Heliebi, Moser, Glasmacher and Brislinger2018). Our evaluation demonstrated that 8 out of 17 investigated placenta-relevant antibodies showed positive immunostaining after pepsin digestion, making the enzymatic method a heatless antigen retrieval alternative for these antibodies. These investigations can help in finding suitable new in vitro models for placental research that are comparable to the in vivo situation.
There is a clear need for a quantitative image-based assessment of antibodies in placental research as considerable knowledge about placental pathologies occurring in the first and second trimesters of pregnancy is mainly gained from in vitro studies using immunostaining assays (Aengenheister et al., Reference Aengenheister, Keevend, Muoth, Schönenberger, Diener, Wick and Buerki-Thurnherr2018; Turco et al., Reference Turco, Gardner, Kay, Hamilton, Prater, Hollinshead, McWhinnie, Esposito, Fernando, Skelton, Reimann, Gribble, Sharkey, Marsh, O'Rahilly, Hemberger, Burton and Moffett2018; Knöfler et al., Reference Knöfler, Haider, Saleh, Pollheimer, Gamage and James2019). In addition, the ability to compare placental cell culture models with the in vivo placenta situation illustrates a major benefit of the present study. Hence, our evaluation serves as a powerful tool for the selection of antibody and pretreatment to be used in implantation and placental research to provide objective and precise results.
Conclusion
The presented evaluation of selected antigen retrieval methods for 17 placenta-relevant antibodies serves as a helpful tool in placentation and implantation diagnostic and research. Automated quantitative image analysis supports the unbiased interpretation of large tissue areas; however, the qualitative assessment of each antibody has to be considered. Hence, our findings provide the potential for further progress in the development of placenta cell culture models and thus the clarification of pregnancy-associated diseases.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S1431927621012630.
Acknowledgment
We thank Dr. Andreas Glasner for recruiting first trimester placental tissue samples for this study. This research was funded in whole by the Austrian Science Fund (FWF) and Christian Doppler Research Association (CDG): PIR7-B28. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author contributions statement
J.F. designed and performed the experiments, analyzed the data, and wrote the manuscript. O.N. designed the experiments and edited the text. C.D. performed the experiments and provided technical assistance. S.G., G.M., I.L.-O., and M.G. provided technical assistance and edited the text. D.B. wrote the manuscript and edited the text. All authors read and approved the final manuscript, and read and met the ICMJE criteria for authorship.
Conflict of interest
The authors declare no conflict of interest.