Healthcare-associated infections (HAIs) are infections acquired as the result of medical care. Reference Collins and Hughes1 The most common HAIs are surgical site infections (SSIs), accounting for >20% of all HAIs. Reference Magill, Edwards and Bamberg2 SSI incidence depends on the type of surgery: in the Netherlands, 1.5% of primary total hip arthroplasties (THAs) and 0.9% of primary total knee arthroplasties (TKAs) are complicated by SSIs, most of which are deep (1.3% and 0.6% respectively). Reference Haque, Sartelli, McKimm and Abu Bakar3,4 This finding is in line with numbers reported in Europe and the United States. 5-Reference Kurtz, Ong, Lau, Mowat and Halpern7 Deep SSIs after THA or TKA are associated with substantial morbidity, longer postoperative hospital stays, and incremental costs. Reference Haque, Sartelli, McKimm and Abu Bakar3,Reference Koek, van der Kooi and Stigter8,Reference Klevens, Edwards and Richards9 Given the aging population, volumes of THA and TKA and numbers of associated SSIs are expected to increase further. Reference Kurtz, Ong, Lau, Mowat and Halpern7,Reference Wolford, Hatfield, Paul, Yi and Slayton10,11
Accurate identification of SSIs through surveillance is essential for targeted implementation and monitoring of interventions to reduce the number of SSIs. Reference Schneeberger, Smits, Zick and Wille12,Reference Haley, Culver and White13 In addition, surveillance data may be used for public reporting and payment mandates. Reference van Mourik, Perencevich, Gastmeier and Bonten14 In most hospitals, surveillance is performed by manual chart review: an infection control practitioner (ICP) reviews electronic health records (EHRs) to determine whether the definition for an SSI is met. This method is costly, time-consuming and labor intensive. Moreover, it is prone to subjectivity, suboptimal interrater reliability, and the “more-you-look-more-you-find” principle. Reference Birgand, Lepelletier and Baron15-Reference Niedner19
The widespread adoption of EHRs facilitates (semi-)automated surveillance using routine care data, thereby reducing workload and improving reliability. SSI surveillance after THA or TKA is particularly suitable for automation because these are high-volume procedures with a low incidence of SSI; hence, the potential gains in efficiency are considerable. In addition, treatment of (possible) SSIs is highly uniform across hospitals, which facilitates algorithmic detection.
As a first step toward semiautomated surveillance of deep SSIs after TKA or THA, a tertiary-care center developed a classification algorithm relying on microbiology results, reinterventions, antibiotic prescriptions, and admission data (Table 1). Reference Sips, Bonten and van Mourik20 This algorithm retrospectively discriminates between patients who have a low or high probability of having developed a deep SSI, and only patients with a high probability undergo manual chart review. Patients classified as low probability are assumed to be free of deep SSI. In a single-hospital setting, this algorithm identified all deep SSIs after THA or TKA (sensitivity of 100%) and resulted in a reduction of 97.3% charts to review. Reference Sips, Bonten and van Mourik20
Table 1. Algorithm specifications

Note. ≥3 of these 4 criteria must be fulfilled to be considered high probability for having deep SSI. Reference Sips, Bonten and van Mourik20 All criteria should be fulfilled within 120 days after the index surgery.
A prerequisite for large-scale implementation of this algorithm is validation in other centers that may differ in EHR systems, patient populations, diagnostic procedures, or clinical practice. Therefore, the main aim of this study was to validate the performance of this algorithm, defined in terms of sensitivity, positive predictive value (PPV), and workload reduction, for semiautomated surveillance to detect deep SSIs after THA or TKA in general hospitals in the Netherlands. A secondary aim was to explore methods for selection of the surveillance population (denominator data).
Methods
Study design
This multicenter retrospective cohort study compares the results of a surveillance algorithm to the results of conventional manual surveillance of deep SSIs following THA and TKA. Manual SSI surveillance, considered the reference standard, was executed according to national definitions and guidelines set out by PREZIES; the Dutch surveillance network for healthcare-associated infections. 21,Reference Verberk, Meijs and Vos22 SSI surveillance includes all patients aged ≥1 year who underwent a primary THA or TKA (so-called index surgery); revision procedures were excluded. SSIs were defined using criteria from the (European) Centers for Disease Control and Prevention, translated and adapted for use in the PREZIES surveillance: organ-space SSIs are reported as deep SSIs. 21,Reference Verberk, Meijs and Vos22 The mandatory follow-up for THA and TKA SSI surveillance is 90 days after the index surgery.
This study was reviewed by the Medical Institutional Review Board of the University Medical Center Utrecht and was considered not to fall under the Medical Research Involving Human Subjects Act. Hence the requirement of an informed consent was waived (reference no. 17-888/C). From all participating hospitals, approval to participate was obtained from the local boards of directors.
Hospitals
We selected 10 hospitals (~14% of all Dutch hospitals) based on their interest in automated surveillance and expected surgical volume and invited them to participate in the study. Hospitals had to meet the following inclusion criteria: (1) recent participation in PREZIES SSI surveillance for THA and TKA according to PREZIES guidelines; (2) availability of at least 2 years of THA and TKA surveillance data after 2012 and data on at least 1,000 surgeries; (3) ability to select the surveillance population (the patients who underwent the index surgery) in electronic hospitals systems; and (4) ability to extract the required routine care data of these patients from the EHR in a structured format to apply the algorithm.
Data collection from electronic health records and algorithm application
Hospitals were requested to automatically select patients who underwent the index surgeries (denominator data) and to extract the following data for these patients from their EHR (Table 1): microbiology results, antibiotic prescriptions, (re)admissions and discharge dates, and subsequent orthopedic surgical procedures. All extracted data were limited to 120 days following the index surgery to enable the algorithm to capture SSIs that developed at the end of the 90-day follow-up period. Data extractions were performed between November 18, 2018, and August 16, 2019. Table S1 provides detailed data specifications.
Analyses
After extraction and cleaning of data, records of patients in the extractions were matched to patients in the reference standard (PREZIES database). If available, matching was performed using a pseudonymized surveillance identification number. Else, matching was performed for the following patient characteristics: date of birth, sex, date of index surgery, date of admission, and type of procedure. For each hospital, the method of automated selection of index surgeries was described as well as the completeness of the surveillance population (denominator) compared to the reference population reported manually to PREZIES. Subsequently, the algorithm was applied, and patients who underwent THA or TKA surgeries were classified as high- or low probability of having had a deep SSI. Patients were classified as high probability for deep SSI according to the algorithm if they met ≥3 of the 4 criteria (Table 1).
For each hospital, the allocation of patients with low or high probability by the algorithm was compared to the outcome (deep SSI) as reported in the reference standard. Subsequently, sensitivity, PPV, and workload reduction (defined as difference between the total number of surgeries in surveillance and the proportion of surgeries requiring manual review after algorithm application) were calculated with corresponding confidence intervals. Reference Clopper and Pearson23 For semiautomated surveillance, we considered sensitivity to be the most important characteristic because any false-positive cases are corrected during subsequent chart review, whereas false-negative cases may remain unnoticed. Analyses were performed using SAS version 9.4 software (SAS Institute, Cary, NC).
Discrepancy analyses and validation of the reference standard
Exploratory discrepancy analyses were performed to evaluate and understand possible underlying causes of misclassification by the algorithm. In addition, for each hospital, an on-site visit took place to validate the conventional surveillance (ie, reference standard PREZIES). This validation was executed by 2 experienced surveillance advisors of PREZIES, and they were blinded for the outcomes both of the reference standard and the algorithm. For the validation of the conventional surveillance, a maximum of 28 records were selected containing all presumed false positives and false negatives, complemented with a random sample of true positives and true negatives. At least 50% of the reported superficial SSIs in the true-negative group were included in the validation sample. The maximum number was selected for logistical reasons and the time capacity of the validation team.
Results
Overall, 4 hospitals met the inclusion criteria and were willing to participate in this study: the Beatrix Hospital in Gorinchem, Haaglanden Medical Center in The Hague (3 locations), Meander Medical Center in Amersfoort and Sint Antonius Hospital in Nieuwegein and Utrecht (3 locations). Hospitals were randomly assigned the letters A, B, C, and D. The remaining hospitals were not able to participate for the following reasons: inability to extract historical data due to a transition of EHR (n = 1); inability to extract microbiology results or antibiotic use from historical data in a structured format (n = 3); no approval of the hospital’s board to share pseudonymized patient data (n = 1) or no capacity of human resources (ICPs, information technology personnel, and data managers) (n = 1).
Completeness of surveillance population
The 4 participating hospitals extracted 9,554 THA and TKA procedures performed between 2012 and 2018 along with data required for application of the algorithm (Table S1, Appendix 1 online). Hospital B used inclusion in conventional surveillance as a selection criterion for the selection of index surgeries and extraction of the data required for the algorithm. These extracted records could be matched using a pseudonymized surveillance identification number, which was also available in the reference standard. By definition, this procedure resulted in a perfect match; hence, no inferences could be made regarding the completeness of the surveillance population when using automated selections, for example, using administrative procedure codes. Hospitals A, C, and D selected their surveillance population automatically using administrative THA and TKA procedure codes. For hospitals A and D, these records were matched by patient characteristics to the reference standard and, for hospital C, by a pseudonymized surveillance identification number. Matching with the PREZIES database revealed a mismatch for 1,128 records that could not be linked to the reference standard. Manual review of a random sample of these records showed that these were mainly revision procedures that were excluded from conventional surveillance. Vice versa, 103 records were in the reference standard but could not be linked to the extractions. Explanations for this mismatch per hospital are described in Table 2.
Table 2. Overview of Data Extractions and Selection of Surveillance Population
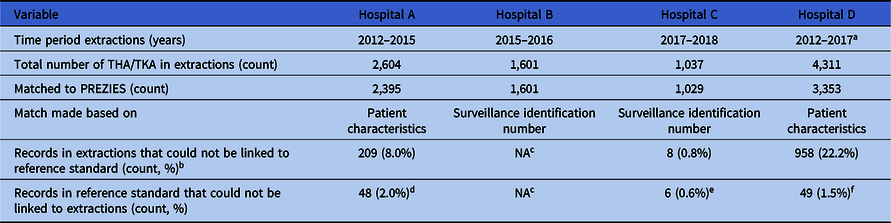
Note. THA, total hip arthroplasty; TKA, total knee arthroplasty; NA, not applicable.
a Until September 1, 2017.
b Manual review of a random sample of these records showed these were mainly revision procedures.
c Exploration of automating selecting surveillance population not applicable as hospital collected data for the extractions based on the selection of the conventional surveillance.
d Reason for mismatch: typo’s and mistakes in the manual data collection.
e Reason for mismatch: all emergency cases for which data was incomplete. Automated extractions therefore not possible.
f Reason for mismatch: a clear cause was not found, although it is suspected data was lost due to the merger of hospitals and their EHR during the study period.
Algorithm performance
In total, 8,378 primary arthroplasty procedures (4,432 THAs and 3,946 TKAs) in 7,659 patients and 95 SSIs (1.1%) were uniquely matched with the reference standard and were available for analysis of algorithm performance (Table 2). The algorithm sensitivity ranged from 93.6% to 100.0% and PPV ranged from 55.8% to 72.2% across hospitals (Table 3). In all hospitals, a workload reduction of ≥98.0% was achieved. In hospitals B and D, 1 and 2 deep SSIs were missed by the algorithm, respectively. Discrepancy analyses revealed that 1 case was reclassified into ‘no deep SSI’, and, hence, was correctly indicated by the algorithm. Of the 2 truly missed cases, 1 case was missed by incomplete microbiology data and the other was missed because of unavailability of data regarding the treating specialty of the readmissions, thereby using ward level for the selection of readmissions. Results and details of false-negative and false-positive cases are provided in Table 4. On-site validation visits found 6 additional deep SSIs, which were missed in the conventional surveillance but were correctly classified as potential SSIs by the algorithm. Other findings of the on-site validation of the reference standard, but not essential for the assessment of the algorithm, were reclassifications of superficial SSIs to no SSI (n = 6), missed superficial SSIs (n = 2), and errors in the determination of the infection date (n = 4).
Table 3. Overview Algorithm Performance per Hospital
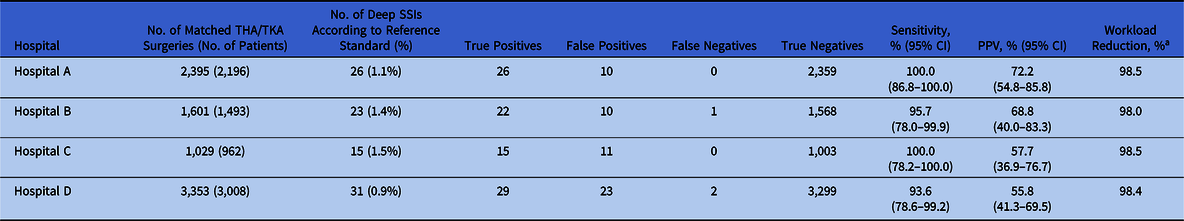
Note. THA, total hip arthroplasty; TKA, total knee arthroplasty; SSI, surgical site infection; PPV, positive predicted value.
a Workload reduction is defined as the proportion of medical records needing manual chart review.
Table 4. Overview of Discrepancy Analysis
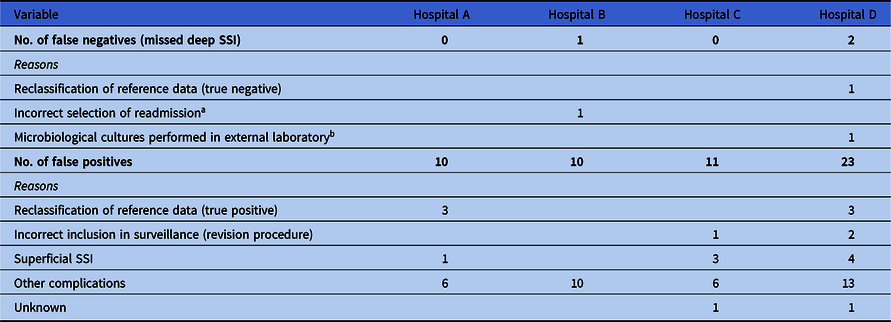
Note. SSI, surgical site infection.
a This hospital used data extractions from a previous electronic health record system, where no information was stored regarding the specialty of the readmission. Selection of readmission was therefore made on ward level, instead of treating specialty. Because this patient was readmitted to another ward because of overcapacity of the orthopedic ward, it was missed by the algorithm.
b Microbiological cultures of this patient were performed in external laboratory and culture results were therefore not available in the in-house laboratory information system from which the data were extracted to apply the algorithm.
Discussion
This study successfully validated a previously developed algorithm for the surveillance of deep SSIs after THA or TKA in 4 hospitals. The algorithm had sensitivity ranging from 93.6% to 100.0% and achieved a workload reduction of 98.0% or more, which is in line with the original study and another international study. Reference Sips, Bonten and van Mourik20,Reference van Rooden, Tacconelli and Pujol24 In total, only 2 SSIs were missed by the algorithm; both were the result of limitations of the use of historical data and can be resolved with the current EHR. Validation of the reference standard revealed 6 additional deep SSIs that were initially missed by conventional surveillance but classified as high probability of SSI by the algorithm; thus, the accuracy of the surveillance improved. For automated selection of the surveillance population (ie, denominator data), hospitals should be able to distinguish primary THAs and TKAs from revisions. Our results provide essential information for successful implementation of semiautomated surveillance for deep SSIs after THA or TKA in Dutch hospitals in the future.
The results of our study reveal some preconditions that require attention when further implementing this algorithm for semiautomated surveillance. First, dialogue between information technology personnel, data management, ICPs and microbiologists is essential to identify the correct sources of data for applying the algorithm. In 2 of 4 hospitals, interim results revealed that data extractions were incomplete due to unawareness in hospitals of the existence of registration codes. This finding demonstrates the importance of validating the completeness and accuracy of data sources required for the implementation of semiautomated surveillance. Reference van Mourik, Perencevich, Gastmeier and Bonten14,Reference Gastmeier and Behnke25 Second, successful validation of this algorithm does not guarantee that widespread implementation can be taken for granted. It appeared that none of the 4 hospitals could perfectly select the surveillance population using structured routine care data such as procedure codes (mismatch ranged from 0.8% to 22.2%). Procedure codes are not developed for the purpose of surveillance but for medico-administrative purposes, and they may contain some misclassification in distinguishing between primary procedures and procedures that should be excluded according to conventional surveillance (eg, revision procedures). Reference Gastmeier and Behnke25 For implementation, improvement of patient selection is considered to increase comparability. Mismatches between the data extractions and reference standard (97 records from the reference standard were not found in the hospitals’ extractions) were partly the result of typing errors in the manual surveillance, hence, they underscore the vulnerability of traditional manual surveillance.
Although superficial SSIs are included within the conventional method of surveillance, this algorithm was developed to detect deep SSIs only. During initial algorithm development superficial SSIs were not taken into account for the following reasons: First, the costs and impact on patient and patient-related outcomes are more detrimental after deep SSIs. In addition, only 20% of all reported SSIs in THAs and TKAs concern superficial SSIs. 4 Third, superficial SSIs are mostly scored by clinical symptoms that are often stored in unstructured data fields (clinical notes) with a wide variety in expressions. Reference Skube, Hu and Arsoniadis26 These data are complex to use in automatization processes and will complicate widespread implementation. Reference Thirukumaran, Zaman and Rubery27 Fourth, the determination of superficial SSIs requires a subjective interpretation of the definition, making them a difficult surveillance target both for manual and automated surveillance.
Previous studies investigating the use of algorithms in SSI surveillance after orthopedic surgeries achieved a low(er) sensitivity, applied rather complex algorithms, or used administrative coding data such as ICD-10 codes for infection. Reference Perdiz, Yokoe, Furtado and Medeiros28-Reference Bolon, Hooper and Stevenson30 Although the use of ICD-10 codes for infection is an easy and straightforward method in some settings, relying solely on administrative data is considered inaccurate. Reference Gastmeier and Behnke25,Reference Curtis, Graves and Birrell31-Reference Goto, Ohl, Schweizer and Perencevich33 In addition, coding practices differ by country, and results cannot be extrapolated. Thirukumaran et al Reference Thirukumaran, Zaman and Rubery27 investigated the use of natural language processing in detecting SSIs. Sensitivity and PPV were extremely high in the center under study; however, the performance in other centers was not investigated, and the proposed method is rather complex to implement on a large scale compared to our method. In contrast, Cho et al Reference Cho, Chung and Choi34 showed a more pragmatic approach in which one algorithm was used to detect SSIs in 38 different procedures, including THAs and TKAs. Although the sensitivity for detecting deep SSIs was 100%, a high number of false positives occurred because of the broad algorithm, resulting in a nonoptimal workload reduction.
Strengths and limitations
The strengths of this study are the multicenter aspect and the use of an algorithm that is relatively simple to apply. All participating hospitals had previously performed conventional surveillance according to a standardized protocol and SSI definitions, enabling optimal comparison and generalizability to the Dutch situation. The algorithm could be successfully applied despite potential differences in clinical and diagnostic practice, as well as the use of different EHRs. Whereas previous studies used complex algorithms and were mostly performed in single tertiary-care centers, this study achieved a near-perfect sensitivity and high workload reduction in small(er) general hospitals, using an algorithm that is likely feasible to implement in these hospitals.
This study has several limitations. First, postdischarge surveillance was limited to patient encounters in the initial hospital. The algorithm will not detect patients who are treated or readmitted in other hospitals; however, this is also the case in conventional surveillance. In the Netherlands, most patients return to the operating hospital in cases with complications such as deep SSIs, especially if they occur within the 90-day follow-up period. Secondly, in this study, we made use of historical data retrieved from the local EHR. Because of shifts in hospital information systems and merger of hospitals, historical data were not accessible for some hospitals, limiting their participation in this retrospective study. Therefore, we have no insight into the feasibility of future large-scale implementation in these hospitals. Lastly, in this study, 1 hospital used the conventional surveillance to identify the surveillance population and to perform electronic data extractions. Therefore, for this hospital, we were unable to adequately evaluate the quality and completeness of the selected surveillance population if they had been using an automated selection procedure.
In conclusion, a previously developed algorithm for semiautomated surveillance of deep SSI after THA and TKA was successfully validated in this multicenter study; a near-perfect sensitivity was reached, with a ≥98% workload reduction. In addition, semiautomated surveillance not only proved to be an efficient method of executing surveillance but also had the potential to capture more true deep SSIs compared to conventional (manual) surveillance approaches. For successful implementation, hospitals should be able to identify the surveillance population using electronically accessible data sources. This study is the first step to broader implementation of semiautomated surveillance in the digital infrastructures of hospitals.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2020.377
Acknowledgments
We thank Meander Sips (UMCU) for helping with the study design and Jan Wille (PREZIES) for performing the on-site validation visits. In addition, we thank the following people for their contribution to the data collection and/or local study coordination: Ada Gigengack (Meander MC), Fabio Bruna (HMC), Desiree Oosterom (HMC), Wilma van Erdewijk (HMC), Saara Vainio (St. Antonius Ziekenhuis), Claudia de Jong (Beatrix Ziekenhuis), Edward Tijssen (Beatrix Ziekenhuis) and Robin van der Vlies (RIVAS Zorggroep).
Financial support
This work was supported by the Regional Healthcare Network Antibiotic Resistance Utrecht with a subsidy of the Dutch Ministry of Health, Welfare and Sport (grant no. 326835).
Conflicts of interest
Drs J.D.M. Verberk, Dr M.S.M. van Mourik, Dr S.R. van Rooden, A.E. Smilde and Dr R.H.R.A. Streefkerk report grants from Regional Healthcare Network Antibiotic Resistance Utrecht for conducting this study. All other authors have nothing to disclose.









