(See introductory commentary by Livorsi et al, pages 186–188.)
Nearly 100 trillion microbes occupying the skin, genitourinary tract, and pulmonary system constitute the human microbiome.Reference Ravel, Blaser and Braun
1
This diverse microbiome consisting of bacteria, fungi, parasites and protozoa plays a significant role in health and disease. Impairment of the gut microbiome is implicated in a diverse array of conditions, both noninfectious and infectious, such as cardiovascular disease,Reference Tang, Wang and Levison
2
Crohn’s disease,Reference Pascal, Pozuelo and Borruel
3
,
Reference Pedamallu, Bhatt and Bullman
4
and Clostridium difficile infection (CDI).Reference Schubert, Rogers and Ring
5
,
Reference Ferreyra, Wu, Hryckowian, Bouley, Weimer and Sonnenburg
6
Improvements in sequencing technology and decreases in sequencing costs have facilitated growth in microbiome research. Projects like the Human Microbiome Project
7
and the European Metagenomics of the Human Intestinal Tract (MetaHIT)Reference Qin, Li and Raes
8
have contributed enormously to our understanding of microbiome complexity and its role in health and disease. Despite this welcome progress, microbiome research is in its infancy, particularly in the context of how the microbiomes influences infection with multidrug resistant organisms (MDRO), including CDI in healthcare settings. Further research aimed at understanding and manipulating the structure and function of the microbiome in human physiology, disease pathogenesis, and infection prevention is necessary.
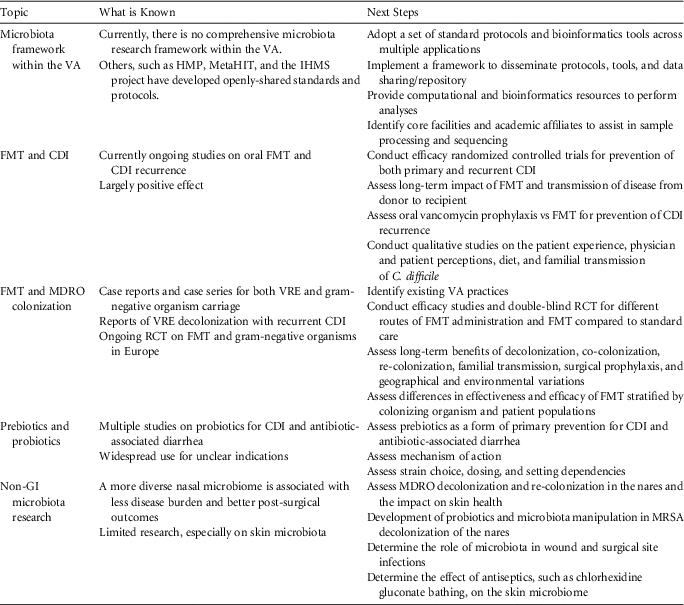
As the largest integrated healthcare system in the United States, serving 8.9 million veterans, the Veterans Health Administration (VHA) system is poised to lead innovative microbiome research in healthcare-associated infections and MDROs. Here, we discuss the proceedings of a multidisciplinary conference on microbiome research in this area across the VHA. This conference highlighted the role of the human microbiome in colonization with MDROs and infection prevention and developed recommendations to guide VHA microbiome research in healthcare-associated infection (HAI) and MDROs (Table 1). The importance of a standardized approach to microbiome research, the role of the gut microbiome with special focus on biotherapeutics, and the role of microbiota at extraintestinal sites are the central focus of these recommendations. While the VHA is uniquely positioned to lead such microbiome research efforts, these proceedings have application beyond the VHA as well.
TABLE 1 Proposed Veterans Health Administration Health Services Research and Development Agenda for Microbiome Research
RECOMMENDATIONS
A Framework for Microbiome Research
A VHA-wide standardized research framework will ensure rigorous, reproducible work and foster collaborations across VHA sites. The need for a set of standards in microbiome research is not unique to the VHA.Reference Ravel, Blaser and Braun
1
,
9
Several methodologies have been applied at all stages of microbiome analysis; however, a gold standard has not emerged. The International Human Microbiome Standards (IHMS) project
10
was established to create a set of standard operating procedures (SOPs), which are crucial to enhancing reproducibility and comparability across microbiome research. Despite the importance of the proposed standards, many labs have not adopted these SOPs.
9
As the VHA advances microbiome research, a standardized research framework must be developed. The recommended components of this framework are shown in Figure 1.
FIGURE 1 Key components of the proposed framework for microbiota research within the Veterans Health Administration (VHA).
To ensure high-quality, reproducible microbiome research, a set of standardized methods should be implemented, such as the IHMS SOPs, which offer clinical definitions, sample collection best practices, and laboratory processes. Novel indices, such as a Microbiome Disruption Index being jointly designed by uBiome and the Centers for Disease Control and Prevention (CDC),
11
may offer promise in tracking the impact of interventions and biotherapeutics on the gut microbiome, can provide early identification of MDRO colonization or infection, and can serve as a method to standardize classification based on set definitions. While a greater understanding of microbiome compositions is required to create standardized classifications, some metrics do currently exist, such as the community states used to classify the vaginal microbiome.Reference Gajer, Brotman and Bai
12
Presently, sample collection procedures vary widely and are dependent on the research question, the anatomic site, the expected biomass, and preferences of the research participant. The cost of the technique and the necessary sample size also influence the sampling procedure. While it is not possible to identify a set of sampling techniques appropriate for all studies, it would be beneficial to curate a set of guidelines and best practices.
Uniformity in laboratory processing methods will optimize the ability to compare results across studies. The method of genome extraction shapes the observed microbial composition due to varying cell lysis techniques.Reference Kuczynski, Lauber and Walters
13
Standardizing methods of genome extraction will require access to core facilities or academic affiliates to carry out sequencing. For VA medical centers without these capabilities, collaboration with a core facility equipped to handle genome extraction is prudent and economical.
Resources for data analysis must be provided to microbiome researchers. Advanced computational and bioinformatics tools are needed to analyze the volume of data produced through next-generation sequencing and shotgun metagenomics studies. Many tools currently exist for analyzing sequencing data, such as Quantitative Insights Into Microbial Ecology (QIIME),Reference Caporaso, Kuczynski and Stombaugh
14
mothur,Reference Schloss, Westcott and Ryabin
15
and UPARSEReference Edgar
16
for targeted amplicon sequencing. However, gaps remain in our understanding of the relative advantages and disadvantages of these methods. After sequencing data are processed, statistical software (R is one such example) must be available for data analysis.
17
In addition to providing access to these programs, the VHA could create a central infrastructure of new and innovative analytic methods to be at the forefront of microbiome research and big data analysis in this context.
Data release and sharing into a repository is essential for microbiome research to ensure reproducibility and validation and allow leveraging of existing or planned studies. Sequencing and metadata should be uploaded to the repository frequently and following normal standards
18
so it can be available to the scientific community. Repository data should be reasonably deidentified, consistent with institutional review board policies.
Study Design Considerations
Much of the published literature on the human microbiome and interventions to treat perturbations of the human microbiome are case studies or uncontrolled case series.Reference Kociolek and Gerding
19
The VHA can advance microbiome research by pursuing longitudinal studies, conducting randomized controlled trials (RCTs) of biotherapeutic interventions, investigating geographic differences in the microbiome, examining patient and provider acceptability of biotherapeutics through qualitative methods, and leveraging the Million Veterans Program as a considerable potential biobank for microbiome data.
Longitudinal studies can elucidate the natural history of changes in the microbiome across time, across the healthcare continuum, and in response to various exposures. Changes in the microbiome should be studied across transitions into and out of the hospital, intensive care units, long-term care facilities, community living centers, and pre- and postoperatively.Reference Halpin and McDonald
20
By comparing groups of patients taking certain medications of interest (eg, antibiotics, metformin), changes to the microbiome related to these exposures can be identified. The influence of disease processes such as diabetes on the microbiome can also be studied in this way.Reference Forslund, Hildebrand and Nielsen
21
The VHA can evaluate the influence of deployment on the microbiome by following the microbiome of military personnel who travel to and from other countries. These longitudinal studies must compare individuals exposed to the variable of interest to those who are not and take care to collect data on other variables that may affect the composition of the microbiome, such as diet, use of antibiotics, and occupation. These variables should be evaluated as risk factors for microbiome perturbations, and adjusted for in analyses.
Randomized controlled trials evaluating the effectiveness of biotherapeutic interventions to limit microbiome perturbation or restore the gut microbiome must be performed. These interventions include, but are not limited to, fecal microbiota transplantation (FMT), probiotics, and nontoxigenic C. difficile. Randomized controlled trials evaluating the use of these biotherapeutics in primary and recurrent CDI and other MDROs should be pursued.Reference Kociolek and Gerding
19
Because these biotherapeutics are organic, studies should account for potential transmission between study subjects, such as nontoxigenic spores transmitted from intervention and control patients.
Both longitudinal studies and RCTs need to be sufficiently powered to detect clinically relevant differences. A recent review by Hanson and WeinstockReference Hanson and Weinstock
22
provides a discussion of methods that can be used for these power calculations.
The VHA should use its strong qualitative and implementation research infrastructure to assess patient and provider views regarding biotherapeutic interventions to alter the microbiome. Patients along the spectrum of health and disease should be surveyed or interviewed to determine attitudes and perceptions toward various types of biotherapeutics and delivery methods.Reference Kociolek and Gerding
19
These studies could reveal differences in risk perception and aversion to biotherapeutics among individuals based on prior disease or carrier status. Similarly, provider perceptions and how they approach conversations regarding these biotherapeutic interventions should be explored.
The VHA system is uniquely poised to perform many of the studies described above. Deployment to developing countries and the ability to connect TriCare data with the VHA Corporate Data Warehouse can provide much of the information needed to perform decades-long longitudinal studies with little added burden to the study subjects. Research to determine geographic variation of microbiomes will be necessary to define a “healthy microbiome” and whether interventions need to be geographically specific, which is important for veterans deployed internationally. The shared electronic medical record across inpatient, outpatient, and community living center care within the VHA provides the opportunity to follow patients during transitions of care. The VHA research funding mechanisms can also encourage investigators to add microbiome analyses to studies of antibiotics, decolonization interventions and new devices.
The Million Veteran Program (MVP) could provide a wealth of information to advance microbiome research. As of August 2016, the MVP had collected blood samples for genetic analysis and has performed baseline and lifestyle (eg, dietary habits, environmental exposures) surveys on more than 500,000 veterans.Reference Gaziano, Concato and Brophy
23
,
24
These data are linked with the electronic medical record to create a mega-biobank to improve understanding of how health is affected by genetics, behaviors, and environmental factors.Reference Gaziano, Concato and Brophy
23
The MVP could also collect stool samples from these Veterans and store them in the mega-biobank, allowing researchers to evaluate how health is affected by the microbiome and how behaviors and environmental factors may be associated with perturbations of the microbiome. By conducting longitudinal observational studies, RCTs and leveraging currently existing resources, the VHA can lead innovative research aimed to fully understand the gut microbiome so it can be leveraged for veteran and civilian health.
INTERVENTIONS TARGETED TOWARD THE MICROBIOME
Fecal Microbiota Transplantation
Gut dysbiosis is the hallmark of both initial and recurrent CDI. Conventional treatments include antibiotics with activity against C. difficile,
Reference Cohen, Gerding and Johnson
25
,
Reference Crook, Walker and Kean
26
but these antibiotics have activity against other gut bacteria, limiting the ability of the microbiota to fully recover following CDI and predisposing patients to recurrence.Reference Chang, Antonopoulos and Kalra
27
Although other factors may also explain why patients have recurrence (eg, low serum antibody response to C. difficile toxins,Reference Kyne, Warny, Qamar and Kelly
28
use of medications such as proton pump inhibitors,Reference Abou Chakra, Pepin, Sirard and Valiquette
29
and the specific strain of C. difficile causing infectionReference Crook, Walker and Kean
26
,
Reference Abou Chakra, Pepin, Sirard and Valiquette
29
), restoration of the gut microbiome through FMT has recently gained acceptance as a safe and effective treatment for CDI. However, the optimal protocol for FMT is unknown; numerous methods of stool preparation, infusion, and recipient and donor preparation have been published.Reference Chapman, Moore and Overbey
30
Variation exists in diluent selected, site of instillation, method of recipient preparation, and donor screening. Fecal microbiota transplantation has been performed in both inpatient and outpatient settings, and self-administration of fecal enema at home has also demonstrated success.Reference Silverman, Davis and Pillai
31
Although there are numerous variables to consider in protocol design, it is encouraging that FMT appears to be effective regardless of the specific protocol.Reference Bakken, Borody and Brandt
32
Studies to evaluate the efficacy and the implementation of FMT for CDI are needed to define best practices.
Several key areas were identified as VHA priorities including efficacy RCTs for both primary and recurrent CDI, long-term assessment of the impact of FMT and transmission of disease from donor to recipient, assessment of oral vancomycin prophylaxis versus FMT for prevention of CDI recurrence, and qualitative studies on the patient experience and provider and patient perceptions.
Primary and severe CDI
Few data are available on the use of FMT for primary, non-recurrent CDI aside from a few case reports. A mathematical model of CDI in an ICU assessed the role of FMT on primary CDIReference Lofgren, Moehring, Anderson, Weber and Fefferman
36
and predicted a decreased median incidence of recurrent CDI in patients treated with FMT for primary CDI. The assumptions of the model require validation and prospective testing to address the role of FMT in primary CDI. In a nonrandomized, open-label, before-and-after prospective study comparing conventional antibiotic treatment for CDI versus early FMT via nasogastric infusion, Lagier et alReference Lagier, Delord and Million
37
found that mortality in the FMT group was significantly less (64.4% vs 18.8%; P<.01). This shift in therapy occurred due to clinical need; their hospital in Marseille developed a ribotype 027 outbreak with a dramatic global mortality rate (50.8%). In an epidemic setting with a high mortality rate, early FMT may be beneficial, but the risks and benefits of FMT in primary CDI in endemic situations have not been well described. Similarly, the evidence for use of FMT in severe CDI consists of published case reports, which suggest efficacy and potential for further study.Reference Neemann, Eichele, Smith, Bociek, Akhtari and Freifeld
38
–
Reference You, Franzos and Holman
41
Patient and provider perceptions of FMT
A commonly cited reason for a limited role of FMT is the aesthetics of treatment. However, few studies exist on patients and provider perceptions regarding FMT. In their 2014 review of FMT for CDI, the Evidence-based Synthesis Program (ESP), a unit of the VA Health Services Research and Development Service, identified no study designed to systematically assess acceptability of FMT among patients. The ESP highlighted these investigations as important next steps in advancing the FMT research agenda.Reference Drekonja, Reich and Gezahegn
42
Zipursky et alReference Zipursky, Sidorsky, Freedman, Sidorsky and Kirkland
43
surveyed 192 outpatients (only 1 had a history of CDI) on their attitudes toward FMT using hypothetical case scenarios. The results were largely positive, with 81% agreeing to FMT for CDI. The need to handle stool and the nasogastric route of administration were the most unappealing aspects of FMT by participants. More respondents (90%; P = .002) agreed to FMT when offered as a pill.
When the same investigators surveyed physician attitudes toward FMT,Reference Zipursky, Sidorsky, Freedman, Sidorsky and Kirkland
44
they found 83 of 135 physicians (65%) in their sample had not offered or referred a patient for FMT. Frequently cited reasons included institutional barriers, concern patients would find it unappealing, and uncertainty regarding indications. Only 8% of physicians believed patients would choose FMT if given the option. As the role of FMT expands, patient and provider perceptions and attitudes will likely evolve to better align.
FMT for MDROs
The hypothesized mechanism of FMT for CDI is that normal colonic microbiota outcompete and competitively exclude C. difficile.Reference van Nood, Vrieze and Nieuwdorp
35
The same principles may apply to other enteric pathogens, such as vancomycin-resistant Enterococcus (VRE). Similar to C. difficile, VRE colonization is induced by microbial dysbiosis. Introduction of commensal microbiota may help eliminate VRE colonization through direct effects such as nutrient depletion or immune-mediated factors. In murine models, FMT is effective at decolonizing VRE.Reference Caballero, Carter and Ke
45
,
Reference Ubeda, Bucci and Caballero
46
In case studies of patients co-colonized with C. difficile and VRE, FMT has resulted in the decolonization of both pathogens.Reference Halpin, de Man and Kraft
47
,
Reference Stripling, Kumar and Baddley
48
However, rigorous evidence of efficacy is lacking, and determining the role of FMT for VRE decolonization requires randomized controlled trials.
Fecal microbiota transplantation may have the potential to reduce colonization with carbapenem-resistant Enterobacteriaceae (CRE). In several small case series, FMT successfully eradicated CRE.Reference Davido, Batista and Michelon
49
,
Reference Lagier, Million, Fournier, Brouqui and Raoult
50
In a recent single-center study of 20 patients receiving FMT for recurrent CDI, DNA microarray analysis identified decreased number and diversity of antibiotic resistance genes, which persisted up to a year following FMT.Reference Millan, Park and Hotte
51
The availability of oral FMT makes it an attractive, relatively low-risk option for eradicating MDRO colonization and warrants further study.
BIOTHERAPEUTICS
The role of prebiotics, probiotics, and synbiotics (mixtures of pre- and probiotics) in the human gastrointestinal (GI) microbiome requires further examination. Prebiotics can be harnessed to enhance community structure and immune responses that confer protection against CDI. For example, fructooligosaccharide and lactulose prebiotics increase resistance to colonization with C. difficile via several mechanisms. The dairy protein glycomacropeptide (GMP) participates in activities related to GI physiology and metabolic disease and beneficially modulates GI microbiota in mice.Reference Sawin, De Wolfe and Aktas
52
When administered in adequate amounts, probiotics confer a health benefit. Several large systematic reviews have demonstrated a reduction in incidence of primary CDI when probiotics were used.Reference Millan, Park and Hotte
51
,
Reference Johnston, Ma and Goldenberg
53
,
Reference Shen, Maw and Tmanova
54
Synbiotics, mixtures of probiotics and prebiotics, confer protection against CDI in mice, reducing viable C. difficile in cecal contents compared to controls. Current limitations to the probiotic literature are that many over-the-counter probiotics have no live organisms, do not have sufficient quantities of microbes, or have no mechanistic basis for their proposed action. These findings may explain the conflicting results from probiotic trials and the gap in our understanding of which probiotics are likely to confer benefit. A recent advance in this area is the use of nontoxigenic C. difficile spores. In a phase 2 RCT, patients who received non-toxigenic C. difficile spores following a resolved episode of CDI had a significant reduction in the risk of recurrence.Reference Gerding, Meyer and Lee
55
The field of biotherapeutics is in its infancy and fundamental biomedical research is needed to understand the potential of these products before widespread implementation and dissemination. Trials of biotherapeutics offer an opportunity to examine GI microbiome changes as a consequence and better understand mechanisms of action.
MICROBIOTA RESEARCH BEYOND THE GASTROINTESTINAL TRACT
The gastrointestinal tract is the most studied human microbiome; however, many other body sites are of clinical relevance to HAI research, particularly the nasal and skin microbiomes.
Methicillin-resistant Staphylococcus aureus (MRSA) has been the focus of nasal microbiome research. MRSA colonization is associated with decreased nasal microbiome diversity and several bacterial species (Streptococcus mitis
Reference Bessesen, Kotter and Wagner
56
and Corynebacterium spp) influence MRSA colonization and virulence.Reference Ramsey, Freire, Gabrilska, Rumbaugh and Lemon
57
,
Reference Uehara, Nakama and Agematsu
58
Studying the effect of therapeutics applied in the nose and longitudinal studies of nasal microbiome changes during hospitalization, antibiotic use and after decolonization are necessary.
Studying the skin microbiome is challenging, but a better understanding of its role is necessary. Constant contact with the environment makes it difficult to distinguish between commensal and transient organisms, and obtaining adequate microbial DNA is challenging given the low biomass of the skin. While skin decolonization with daily or presurgical use of chlorhexidine to reduce infection is becoming more widespread, the impact of antiseptics on long-term skin health and microbial composition is unknown. In addition, the role of the skin microbiome in wound and surgical site infections and the impact of the hand microbiome of healthcare workers in MDRO transmission warrants further study.
CONCLUSIONS
The meeting of a multidisciplinary group of researchers focused on infection prevention and microbiota research identified several key recommendations for the future of microbiota research within the VHA. First, a unified framework including standardized methodologies, access to bioinformatics and computational resources, and provision of a data repository will allow high-quality, reproducible work and collaboration across multiple VHA sites. Incorporating microbiome data into data collection, particularly in infection prevention, will provide the ability to evaluate relationships of the microbiome to lifestyle and clinical factors. Longitudinal studies will elucidate changes in the human microbiome over time and across transitions of care. High-quality RCTs examining the efficacy of various approaches to FMT and qualitative studies exploring patient and provider preferences will clarify the role of FMT in restoration of health and treatment of disease. Additional biotherapeutics should be examined, such as prebiotics, probiotics and synbiotics. Finally, the role of extraintestinal microbiomes, particularly the nose and skin, in health and disease should be studied further.
Given the large role the VHA plays in the health of veterans and civilians and its strong research infrastructure, the VHA is well positioned to lead the next wave of discovery in human microbiome research.






(See introductory commentary by Livorsi et al, pages 186–188.)
Nearly 100 trillion microbes occupying the skin, genitourinary tract, and pulmonary system constitute the human microbiome.Reference Ravel, Blaser and Braun 1 This diverse microbiome consisting of bacteria, fungi, parasites and protozoa plays a significant role in health and disease. Impairment of the gut microbiome is implicated in a diverse array of conditions, both noninfectious and infectious, such as cardiovascular disease,Reference Tang, Wang and Levison 2 Crohn’s disease,Reference Pascal, Pozuelo and Borruel 3 , Reference Pedamallu, Bhatt and Bullman 4 and Clostridium difficile infection (CDI).Reference Schubert, Rogers and Ring 5 , Reference Ferreyra, Wu, Hryckowian, Bouley, Weimer and Sonnenburg 6
Improvements in sequencing technology and decreases in sequencing costs have facilitated growth in microbiome research. Projects like the Human Microbiome Project 7 and the European Metagenomics of the Human Intestinal Tract (MetaHIT)Reference Qin, Li and Raes 8 have contributed enormously to our understanding of microbiome complexity and its role in health and disease. Despite this welcome progress, microbiome research is in its infancy, particularly in the context of how the microbiomes influences infection with multidrug resistant organisms (MDRO), including CDI in healthcare settings. Further research aimed at understanding and manipulating the structure and function of the microbiome in human physiology, disease pathogenesis, and infection prevention is necessary.
As the largest integrated healthcare system in the United States, serving 8.9 million veterans, the Veterans Health Administration (VHA) system is poised to lead innovative microbiome research in healthcare-associated infections and MDROs. Here, we discuss the proceedings of a multidisciplinary conference on microbiome research in this area across the VHA. This conference highlighted the role of the human microbiome in colonization with MDROs and infection prevention and developed recommendations to guide VHA microbiome research in healthcare-associated infection (HAI) and MDROs (Table 1). The importance of a standardized approach to microbiome research, the role of the gut microbiome with special focus on biotherapeutics, and the role of microbiota at extraintestinal sites are the central focus of these recommendations. While the VHA is uniquely positioned to lead such microbiome research efforts, these proceedings have application beyond the VHA as well.
TABLE 1 Proposed Veterans Health Administration Health Services Research and Development Agenda for Microbiome Research
NOTE. VA, Veterans Affairs; HMP, Human Microbiome Project; IHMS, International Human Microbiome Standards; CDI, Clostridium difficile infection; FMT, fecal microbiota transplantation; MDRO, multidrug-resistant organism; VRE, vancomycin-resistant Enterococcus; RCT, randomized controlled trial; GI, gastrointestinal; MRSA, methicillin-resistant Staphylococcus aureus.
RECOMMENDATIONS
A Framework for Microbiome Research
A VHA-wide standardized research framework will ensure rigorous, reproducible work and foster collaborations across VHA sites. The need for a set of standards in microbiome research is not unique to the VHA.Reference Ravel, Blaser and Braun 1 , 9 Several methodologies have been applied at all stages of microbiome analysis; however, a gold standard has not emerged. The International Human Microbiome Standards (IHMS) project 10 was established to create a set of standard operating procedures (SOPs), which are crucial to enhancing reproducibility and comparability across microbiome research. Despite the importance of the proposed standards, many labs have not adopted these SOPs. 9 As the VHA advances microbiome research, a standardized research framework must be developed. The recommended components of this framework are shown in Figure 1.
FIGURE 1 Key components of the proposed framework for microbiota research within the Veterans Health Administration (VHA).
To ensure high-quality, reproducible microbiome research, a set of standardized methods should be implemented, such as the IHMS SOPs, which offer clinical definitions, sample collection best practices, and laboratory processes. Novel indices, such as a Microbiome Disruption Index being jointly designed by uBiome and the Centers for Disease Control and Prevention (CDC), 11 may offer promise in tracking the impact of interventions and biotherapeutics on the gut microbiome, can provide early identification of MDRO colonization or infection, and can serve as a method to standardize classification based on set definitions. While a greater understanding of microbiome compositions is required to create standardized classifications, some metrics do currently exist, such as the community states used to classify the vaginal microbiome.Reference Gajer, Brotman and Bai 12
Presently, sample collection procedures vary widely and are dependent on the research question, the anatomic site, the expected biomass, and preferences of the research participant. The cost of the technique and the necessary sample size also influence the sampling procedure. While it is not possible to identify a set of sampling techniques appropriate for all studies, it would be beneficial to curate a set of guidelines and best practices.
Uniformity in laboratory processing methods will optimize the ability to compare results across studies. The method of genome extraction shapes the observed microbial composition due to varying cell lysis techniques.Reference Kuczynski, Lauber and Walters 13 Standardizing methods of genome extraction will require access to core facilities or academic affiliates to carry out sequencing. For VA medical centers without these capabilities, collaboration with a core facility equipped to handle genome extraction is prudent and economical.
Resources for data analysis must be provided to microbiome researchers. Advanced computational and bioinformatics tools are needed to analyze the volume of data produced through next-generation sequencing and shotgun metagenomics studies. Many tools currently exist for analyzing sequencing data, such as Quantitative Insights Into Microbial Ecology (QIIME),Reference Caporaso, Kuczynski and Stombaugh 14 mothur,Reference Schloss, Westcott and Ryabin 15 and UPARSEReference Edgar 16 for targeted amplicon sequencing. However, gaps remain in our understanding of the relative advantages and disadvantages of these methods. After sequencing data are processed, statistical software (R is one such example) must be available for data analysis. 17 In addition to providing access to these programs, the VHA could create a central infrastructure of new and innovative analytic methods to be at the forefront of microbiome research and big data analysis in this context.
Data release and sharing into a repository is essential for microbiome research to ensure reproducibility and validation and allow leveraging of existing or planned studies. Sequencing and metadata should be uploaded to the repository frequently and following normal standards 18 so it can be available to the scientific community. Repository data should be reasonably deidentified, consistent with institutional review board policies.
Study Design Considerations
Much of the published literature on the human microbiome and interventions to treat perturbations of the human microbiome are case studies or uncontrolled case series.Reference Kociolek and Gerding 19 The VHA can advance microbiome research by pursuing longitudinal studies, conducting randomized controlled trials (RCTs) of biotherapeutic interventions, investigating geographic differences in the microbiome, examining patient and provider acceptability of biotherapeutics through qualitative methods, and leveraging the Million Veterans Program as a considerable potential biobank for microbiome data.
Longitudinal studies can elucidate the natural history of changes in the microbiome across time, across the healthcare continuum, and in response to various exposures. Changes in the microbiome should be studied across transitions into and out of the hospital, intensive care units, long-term care facilities, community living centers, and pre- and postoperatively.Reference Halpin and McDonald 20 By comparing groups of patients taking certain medications of interest (eg, antibiotics, metformin), changes to the microbiome related to these exposures can be identified. The influence of disease processes such as diabetes on the microbiome can also be studied in this way.Reference Forslund, Hildebrand and Nielsen 21 The VHA can evaluate the influence of deployment on the microbiome by following the microbiome of military personnel who travel to and from other countries. These longitudinal studies must compare individuals exposed to the variable of interest to those who are not and take care to collect data on other variables that may affect the composition of the microbiome, such as diet, use of antibiotics, and occupation. These variables should be evaluated as risk factors for microbiome perturbations, and adjusted for in analyses.
Randomized controlled trials evaluating the effectiveness of biotherapeutic interventions to limit microbiome perturbation or restore the gut microbiome must be performed. These interventions include, but are not limited to, fecal microbiota transplantation (FMT), probiotics, and nontoxigenic C. difficile. Randomized controlled trials evaluating the use of these biotherapeutics in primary and recurrent CDI and other MDROs should be pursued.Reference Kociolek and Gerding 19 Because these biotherapeutics are organic, studies should account for potential transmission between study subjects, such as nontoxigenic spores transmitted from intervention and control patients.
Both longitudinal studies and RCTs need to be sufficiently powered to detect clinically relevant differences. A recent review by Hanson and WeinstockReference Hanson and Weinstock 22 provides a discussion of methods that can be used for these power calculations.
The VHA should use its strong qualitative and implementation research infrastructure to assess patient and provider views regarding biotherapeutic interventions to alter the microbiome. Patients along the spectrum of health and disease should be surveyed or interviewed to determine attitudes and perceptions toward various types of biotherapeutics and delivery methods.Reference Kociolek and Gerding 19 These studies could reveal differences in risk perception and aversion to biotherapeutics among individuals based on prior disease or carrier status. Similarly, provider perceptions and how they approach conversations regarding these biotherapeutic interventions should be explored.
The VHA system is uniquely poised to perform many of the studies described above. Deployment to developing countries and the ability to connect TriCare data with the VHA Corporate Data Warehouse can provide much of the information needed to perform decades-long longitudinal studies with little added burden to the study subjects. Research to determine geographic variation of microbiomes will be necessary to define a “healthy microbiome” and whether interventions need to be geographically specific, which is important for veterans deployed internationally. The shared electronic medical record across inpatient, outpatient, and community living center care within the VHA provides the opportunity to follow patients during transitions of care. The VHA research funding mechanisms can also encourage investigators to add microbiome analyses to studies of antibiotics, decolonization interventions and new devices.
The Million Veteran Program (MVP) could provide a wealth of information to advance microbiome research. As of August 2016, the MVP had collected blood samples for genetic analysis and has performed baseline and lifestyle (eg, dietary habits, environmental exposures) surveys on more than 500,000 veterans.Reference Gaziano, Concato and Brophy 23 , 24 These data are linked with the electronic medical record to create a mega-biobank to improve understanding of how health is affected by genetics, behaviors, and environmental factors.Reference Gaziano, Concato and Brophy 23 The MVP could also collect stool samples from these Veterans and store them in the mega-biobank, allowing researchers to evaluate how health is affected by the microbiome and how behaviors and environmental factors may be associated with perturbations of the microbiome. By conducting longitudinal observational studies, RCTs and leveraging currently existing resources, the VHA can lead innovative research aimed to fully understand the gut microbiome so it can be leveraged for veteran and civilian health.
INTERVENTIONS TARGETED TOWARD THE MICROBIOME
Fecal Microbiota Transplantation
Gut dysbiosis is the hallmark of both initial and recurrent CDI. Conventional treatments include antibiotics with activity against C. difficile, Reference Cohen, Gerding and Johnson 25 , Reference Crook, Walker and Kean 26 but these antibiotics have activity against other gut bacteria, limiting the ability of the microbiota to fully recover following CDI and predisposing patients to recurrence.Reference Chang, Antonopoulos and Kalra 27 Although other factors may also explain why patients have recurrence (eg, low serum antibody response to C. difficile toxins,Reference Kyne, Warny, Qamar and Kelly 28 use of medications such as proton pump inhibitors,Reference Abou Chakra, Pepin, Sirard and Valiquette 29 and the specific strain of C. difficile causing infectionReference Crook, Walker and Kean 26 , Reference Abou Chakra, Pepin, Sirard and Valiquette 29 ), restoration of the gut microbiome through FMT has recently gained acceptance as a safe and effective treatment for CDI. However, the optimal protocol for FMT is unknown; numerous methods of stool preparation, infusion, and recipient and donor preparation have been published.Reference Chapman, Moore and Overbey 30 Variation exists in diluent selected, site of instillation, method of recipient preparation, and donor screening. Fecal microbiota transplantation has been performed in both inpatient and outpatient settings, and self-administration of fecal enema at home has also demonstrated success.Reference Silverman, Davis and Pillai 31 Although there are numerous variables to consider in protocol design, it is encouraging that FMT appears to be effective regardless of the specific protocol.Reference Bakken, Borody and Brandt 32 Studies to evaluate the efficacy and the implementation of FMT for CDI are needed to define best practices.
Several key areas were identified as VHA priorities including efficacy RCTs for both primary and recurrent CDI, long-term assessment of the impact of FMT and transmission of disease from donor to recipient, assessment of oral vancomycin prophylaxis versus FMT for prevention of CDI recurrence, and qualitative studies on the patient experience and provider and patient perceptions.
Recurrent CDI
The clinical evidence for FMT is most robust for recurrent CDI, and 3 RCTs in this field have helped to clarify the efficacy of FMT.Reference Youngster, Russell, Pindar, Ziv-Baran, Sauk and Hohmann 33 – Reference van Nood, Vrieze and Nieuwdorp 35 Each found rates of cure greater than 70%–90% following FMT; however, sample size, methodologic limitations, need for multiple FMT in several patients and limited duration of follow-up in these studies means there remains a need for high-quality prospective trials.
Primary and severe CDI
Few data are available on the use of FMT for primary, non-recurrent CDI aside from a few case reports. A mathematical model of CDI in an ICU assessed the role of FMT on primary CDIReference Lofgren, Moehring, Anderson, Weber and Fefferman 36 and predicted a decreased median incidence of recurrent CDI in patients treated with FMT for primary CDI. The assumptions of the model require validation and prospective testing to address the role of FMT in primary CDI. In a nonrandomized, open-label, before-and-after prospective study comparing conventional antibiotic treatment for CDI versus early FMT via nasogastric infusion, Lagier et alReference Lagier, Delord and Million 37 found that mortality in the FMT group was significantly less (64.4% vs 18.8%; P<.01). This shift in therapy occurred due to clinical need; their hospital in Marseille developed a ribotype 027 outbreak with a dramatic global mortality rate (50.8%). In an epidemic setting with a high mortality rate, early FMT may be beneficial, but the risks and benefits of FMT in primary CDI in endemic situations have not been well described. Similarly, the evidence for use of FMT in severe CDI consists of published case reports, which suggest efficacy and potential for further study.Reference Neemann, Eichele, Smith, Bociek, Akhtari and Freifeld 38 – Reference You, Franzos and Holman 41
Patient and provider perceptions of FMT
A commonly cited reason for a limited role of FMT is the aesthetics of treatment. However, few studies exist on patients and provider perceptions regarding FMT. In their 2014 review of FMT for CDI, the Evidence-based Synthesis Program (ESP), a unit of the VA Health Services Research and Development Service, identified no study designed to systematically assess acceptability of FMT among patients. The ESP highlighted these investigations as important next steps in advancing the FMT research agenda.Reference Drekonja, Reich and Gezahegn 42
Zipursky et alReference Zipursky, Sidorsky, Freedman, Sidorsky and Kirkland 43 surveyed 192 outpatients (only 1 had a history of CDI) on their attitudes toward FMT using hypothetical case scenarios. The results were largely positive, with 81% agreeing to FMT for CDI. The need to handle stool and the nasogastric route of administration were the most unappealing aspects of FMT by participants. More respondents (90%; P = .002) agreed to FMT when offered as a pill.
When the same investigators surveyed physician attitudes toward FMT,Reference Zipursky, Sidorsky, Freedman, Sidorsky and Kirkland 44 they found 83 of 135 physicians (65%) in their sample had not offered or referred a patient for FMT. Frequently cited reasons included institutional barriers, concern patients would find it unappealing, and uncertainty regarding indications. Only 8% of physicians believed patients would choose FMT if given the option. As the role of FMT expands, patient and provider perceptions and attitudes will likely evolve to better align.
FMT for MDROs
The hypothesized mechanism of FMT for CDI is that normal colonic microbiota outcompete and competitively exclude C. difficile.Reference van Nood, Vrieze and Nieuwdorp 35 The same principles may apply to other enteric pathogens, such as vancomycin-resistant Enterococcus (VRE). Similar to C. difficile, VRE colonization is induced by microbial dysbiosis. Introduction of commensal microbiota may help eliminate VRE colonization through direct effects such as nutrient depletion or immune-mediated factors. In murine models, FMT is effective at decolonizing VRE.Reference Caballero, Carter and Ke 45 , Reference Ubeda, Bucci and Caballero 46 In case studies of patients co-colonized with C. difficile and VRE, FMT has resulted in the decolonization of both pathogens.Reference Halpin, de Man and Kraft 47 , Reference Stripling, Kumar and Baddley 48 However, rigorous evidence of efficacy is lacking, and determining the role of FMT for VRE decolonization requires randomized controlled trials.
Fecal microbiota transplantation may have the potential to reduce colonization with carbapenem-resistant Enterobacteriaceae (CRE). In several small case series, FMT successfully eradicated CRE.Reference Davido, Batista and Michelon 49 , Reference Lagier, Million, Fournier, Brouqui and Raoult 50 In a recent single-center study of 20 patients receiving FMT for recurrent CDI, DNA microarray analysis identified decreased number and diversity of antibiotic resistance genes, which persisted up to a year following FMT.Reference Millan, Park and Hotte 51 The availability of oral FMT makes it an attractive, relatively low-risk option for eradicating MDRO colonization and warrants further study.
BIOTHERAPEUTICS
The role of prebiotics, probiotics, and synbiotics (mixtures of pre- and probiotics) in the human gastrointestinal (GI) microbiome requires further examination. Prebiotics can be harnessed to enhance community structure and immune responses that confer protection against CDI. For example, fructooligosaccharide and lactulose prebiotics increase resistance to colonization with C. difficile via several mechanisms. The dairy protein glycomacropeptide (GMP) participates in activities related to GI physiology and metabolic disease and beneficially modulates GI microbiota in mice.Reference Sawin, De Wolfe and Aktas 52
When administered in adequate amounts, probiotics confer a health benefit. Several large systematic reviews have demonstrated a reduction in incidence of primary CDI when probiotics were used.Reference Millan, Park and Hotte 51 , Reference Johnston, Ma and Goldenberg 53 , Reference Shen, Maw and Tmanova 54 Synbiotics, mixtures of probiotics and prebiotics, confer protection against CDI in mice, reducing viable C. difficile in cecal contents compared to controls. Current limitations to the probiotic literature are that many over-the-counter probiotics have no live organisms, do not have sufficient quantities of microbes, or have no mechanistic basis for their proposed action. These findings may explain the conflicting results from probiotic trials and the gap in our understanding of which probiotics are likely to confer benefit. A recent advance in this area is the use of nontoxigenic C. difficile spores. In a phase 2 RCT, patients who received non-toxigenic C. difficile spores following a resolved episode of CDI had a significant reduction in the risk of recurrence.Reference Gerding, Meyer and Lee 55
The field of biotherapeutics is in its infancy and fundamental biomedical research is needed to understand the potential of these products before widespread implementation and dissemination. Trials of biotherapeutics offer an opportunity to examine GI microbiome changes as a consequence and better understand mechanisms of action.
MICROBIOTA RESEARCH BEYOND THE GASTROINTESTINAL TRACT
The gastrointestinal tract is the most studied human microbiome; however, many other body sites are of clinical relevance to HAI research, particularly the nasal and skin microbiomes.
Methicillin-resistant Staphylococcus aureus (MRSA) has been the focus of nasal microbiome research. MRSA colonization is associated with decreased nasal microbiome diversity and several bacterial species (Streptococcus mitis Reference Bessesen, Kotter and Wagner 56 and Corynebacterium spp) influence MRSA colonization and virulence.Reference Ramsey, Freire, Gabrilska, Rumbaugh and Lemon 57 , Reference Uehara, Nakama and Agematsu 58 Studying the effect of therapeutics applied in the nose and longitudinal studies of nasal microbiome changes during hospitalization, antibiotic use and after decolonization are necessary.
Studying the skin microbiome is challenging, but a better understanding of its role is necessary. Constant contact with the environment makes it difficult to distinguish between commensal and transient organisms, and obtaining adequate microbial DNA is challenging given the low biomass of the skin. While skin decolonization with daily or presurgical use of chlorhexidine to reduce infection is becoming more widespread, the impact of antiseptics on long-term skin health and microbial composition is unknown. In addition, the role of the skin microbiome in wound and surgical site infections and the impact of the hand microbiome of healthcare workers in MDRO transmission warrants further study.
CONCLUSIONS
The meeting of a multidisciplinary group of researchers focused on infection prevention and microbiota research identified several key recommendations for the future of microbiota research within the VHA. First, a unified framework including standardized methodologies, access to bioinformatics and computational resources, and provision of a data repository will allow high-quality, reproducible work and collaboration across multiple VHA sites. Incorporating microbiome data into data collection, particularly in infection prevention, will provide the ability to evaluate relationships of the microbiome to lifestyle and clinical factors. Longitudinal studies will elucidate changes in the human microbiome over time and across transitions of care. High-quality RCTs examining the efficacy of various approaches to FMT and qualitative studies exploring patient and provider preferences will clarify the role of FMT in restoration of health and treatment of disease. Additional biotherapeutics should be examined, such as prebiotics, probiotics and synbiotics. Finally, the role of extraintestinal microbiomes, particularly the nose and skin, in health and disease should be studied further.
Given the large role the VHA plays in the health of veterans and civilians and its strong research infrastructure, the VHA is well positioned to lead the next wave of discovery in human microbiome research.
ACKNOWLEDGMENTS
Financial support: This work was supported in part by funding from the VA Health Services Research and Development (HSR&D) Service Center of Innovation (COIN) conference supplement (“Setting the Clinical Research Agenda for MDROs in VA,” grant no. CIN 13-412) and by the VA Quality Enhancement Research Initiative CARRIAGE Program (grant no. IP1 HX001993-01A1). The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the US government. DNG holds patents for treatment and prevention of CDI, is a member of Advisory Boards for Merck, Summit, Actelion, Rebiotix and DaVolterra and is a consultant for Sanofi Pasteur, MGB Pharma and Pfizer.