INTRODUCTION
Rotaviruses, family Reoviridae, are classified into seven groups (A–G) on basis of antigenicity of the inner capsid VP6 protein [Reference Kapikian, Hoshino, Chanock, Knipe, Howley, Griffin, Lamb, Martin, Roizman and Straus1]. Among them, group A rotaviruses (GAR) are regarded as a major cause of acute diarrhoea in young of humans, animals, and birds [Reference Kapikian, Hoshino, Chanock, Knipe, Howley, Griffin, Lamb, Martin, Roizman and Straus1]. Unlike GAR, group B rotaviruses (GBR) have been primarily associated with adult diarrhoea [Reference Kobayashi, Naik, Ahmed and Kobayashi2, Reference Sen, Raghunath and Nayak3], and are thought to be a rare cause of gastroenteritis in human infants, calves, lambs, pigs, and rats [Reference Kobayashi, Naik, Ahmed and Kobayashi2–Reference Vonderfecht, Lindsay and Eiden10]. In cattle, limited surveillance studies have reported the association of bovine GBR in outbreaks or sporadic cases of diarrhoeal diseases from Japan [Reference Tsunemitsu8, Reference Tsunemitsu11, Reference Hayashi12], United Kingdom [Reference Bridger13], United States [Reference Chang6, Reference Chinsangaram7, Reference Saif14], and India [Reference Barman15]. Moreover, little information is available on the genetic make-up of bovine GBR strains, and only gene segment 9 [VP7 encoding gene, 811–816 nucleotides (nt) long depending upon the strain, with an intact open reading frame from nt 15 to nt 758], encoding the outer capsid VP7 protein (equivalent of group A neutralizing antigen VP7), has been sequenced for all the representative bovine GBR strains (ATI, WD653, Mebus, Nemuro and Kolkata strains) [Reference Chang6, Reference Tsunemitsu11, Reference Barman15]. Among other genes, the VP6 gene of Nemuro strain [Reference Tsunemitsu8] and NSP5 gene of Kolkata strains [Reference Barman15] have been sequenced. The three Indian GBR strains (referred to as the Kolkata strains: DB101, DB176, DB180) were detected from calves in Kolkata city, Eastern India, and exhibited a novel VP7 genotype [Reference Barman15]. In the present study, we report a significant increase in prevalence of the ‘Kolkata strain’-like novel GBR strains from calves (aged <1 month) with diarrhoea, and provide evidence for interstate spread of bovine GBR infection.
MATERIALS AND METHODS
Sampling
In between December 2003 and March 2005, 260 faecal samples were collected from calves (aged <6 months) with diarrhoea from a cattle market located in Kolkata, capital city of the state of West Bengal, Eastern India. The cattle market sold calves transported overnight by road in trucks from the neighbouring states of West Bengal. Faecal samples were collected from diarrhoeic calves following their arrival in the cattle market in the morning, and before they were sold within a few hours. After the arrival of the calves, the investigators searched the market for the next 1–2 h, identifying calves with diarrhoea by clinical symptoms (defecating loose stool and exhibiting signs of dehydration) and interviewing the owner of the animal. Subsequently, samples were collected from these calves by the per-rectal route, transferred to sterile vials, transported to the laboratory, and stored under refrigeration. To trace the origin of infection, case histories were collected from the farmers accompanying the calves to the market and were recorded in data sheets. On an average, 10–12 calves arrived each day in the cattle market, and of them, from none to 2–5 calves were diarrhoeic. The frequency of collection was none to 2–5 samples per day, once or twice a week, during the entire surveillance period of December 2003 to March 2005.
Detection of rotaviruses
For screening of samples for the presence of rotaviruses, virus RNA was extracted by the standard phenol chloroform extraction method, electrophoresis of RNA was carried out in 10% polyacrylamide gels, and the gels were visualized by the silver staining method as previously described [Reference Herring16].
Reverse transcription–polymerase chain reaction (RT–PCR)
Extraction of virus double-stranded RNA for RT–PCR assays was carried out using the commercially available column-based Nucleospin RNA extraction kit (Macherey Nagel, GmbH & Co., Duren, Germany) according to the manufacturer's instructions. Primer pairs MEV7-1 (5′-GGA AAT AAT CAG AGA TGC CGT T-3′, nt 1–22) and MEV7-3 (5′-CTA CTC GTT TGG CTC CCT CC-3′, nt 795–776) were used to amplify the VP7 encoding gene of bovine GBR strains by RT–PCR assays as previously described [Reference Barman15]. Similarly, amplification of the NSP5 gene was achieved using primer pairs BBR11-3F (5′-GGA ATA AAA GAG ACA GGT AG-3′, nt 1–20) and BBR11-3R (5′-GGG TAT TAT TCC AGC ACT AA-3′, nt 625–606) as previously described [Reference Barman15].
Sequencing and sequence analyses
Partial GBR VP7 and NSP5 sequences were generated using the forward primer (MEV7-1 for VP7 and BBR11-3F for NSP5) in an ABI Prism 3100 Genetic Analyzer (PE Applied Biosystems, Foster City, CA, USA). The entire process (RNA extraction to sequencing) was repeated three times for each gene sequenced to rule out any ambiguities. Sequence analyses were performed as previously described [Reference Barman15, Reference Ghosh17, Reference Varghese18]. In brief, homology search for cognate sequences was performed using blast search software, pair-wise amino acid (aa) alignments were carried out using the lalign program with the global alignment option (http://www.ch.embnet.org/software/LALIGNform.html), and the phylogenetic tree was constructed by the neighbour-joining method [Reference Saitou and Nei19] (random number generator seed of 111 and 1000 bootstrap trials) using the Clustal X (version 1.8) software program, and drawn with TreeView (Version 1.6.6).
Nucleotide sequence accession numbers
The VP7 gene sequences of the 28 GBR (RUBV) strains detected during the study period were deposited in the GenBank database, and assigned accession numbers DQ531532–DQ531559. Similarly, the NSP5 gene nucleotide sequences of strains RUBV79, RUBV99, RUBV102, RUBV106, RUBV137, RUBV282 and RUBV306 were assigned accession numbers DQ98761–DQ987867. Accession numbers of nucleotide sequences of GBR strains used from the database for comparative analyses were U84472, U84141, U84473, AB016818, AY158155, AF531910, AF529214, D00911, AF184083, AY238385, AY539856, M33872 for the VP7 gene of ATI, WD653, Mebus, Nemuro, DB101, DB176, DB180, IDIR, CAL, Bang373, WH-1, ADRV strains, and AF079158, AY347930, AY347929, AY347928, M34380, AY238386, AY238394, D00912 for the NSP5 gene of KB63, DB101, DB176, DB180, ADRV, CAL, Bang373, IDIR strains respectively.
RESULTS
Incidence
Out of 260 samples collected from diarrhoeic calves in the cattle market in Kolkata city, West Bengal, Eastern India, 113 (43·4%) and 34 (13·0%) were positive for GAR and GBR respectively. All the 34 GBR-positive samples exhibited the characteristic bovine GBR ‘4-2-2-3’ RNA migration patterns in polyacrylamide gels (Fig. 1). The GBR-positive cases were from calves aged <1 month and were detected non-consecutively. The infection was associated with sporadic cases of diarrhoea and was detected throughout the year (Table 1). Case histories collected from the farmers accompanying the calves to the cattle market traced the GBR-infected calves to districts in the neighbouring states of West Bengal (Tables 1 and 2, Fig. 2).

Fig. 1. Migration pattern of rotavirus double-stranded RNA in 10% polyacrylamide gels. Group B rotavirus Kolkata strain ‘DB176’ (lane 1) and G6P[Reference Tsunemitsu11] group A rotavirus strain ‘RUBV10’ (detected and characterized in our laboratory) (lane 7) have been used as positive controls. The other representative group B rotavirus strains detected during the course of the present study with their names have been shown in lanes 2–6.

Fig. 2. Map of India highlighting the cattle market in Kolkata city, capital of the state of West Bengal, and the four districts (■) [Dumka (State of Jharkhand), Chhapra and Purnia (State of Bihar) and Gorakhpur (State of Uttar Pradesh)] in neighbouring states of West Bengal from where the calves were being transported to the cattle market. The approximate distance (in km) between the Kolkata cattle market and the four districts have been indicated. The map is not to scale.
Table 1. Details of group B rotavirus strains detected from calves with diarrhoea in the Kolkata cattle market, Kolkata city, state of West Bengal, India between December 2003 and March 2005

nt, Nucleotides.
* As revealed by case histories collected from farmers accompanying calves to the market.
Table 2. District wise distribution of calves positive for group B rotaviruses from both the surveillance studies conducted in the cattle market in Kolkata city, capital of state of West Bengal, Eastern India
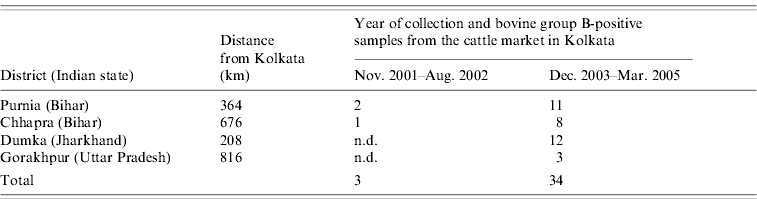
n.d., Not detected.
RT–PCR
Out of the 34 samples positive for GBR by RNA electrophoresis in polyacrylamide gels, 28 were available in sufficient quantity for RT–PCR-based amplification of the VP7 encoding gene. All the 28 strains (designated as RUBV strains) exhibited the expected 795 bp VP7 amplicon with primer pairs MEV7-1 and MEV7-3 [Reference Barman15], and were selected for sequencing. Moreover, to further establish the genetic relatedness between the RUBV and Kolkata strains, the NSP5 gene (625 nt long) of seven randomly selected RUBV strains was amplified using primer pairs BBR11-3F and BBR11-3R [Reference Barman15], and the 625 bp amplicons were sequenced.
Sequencing
Partial VP7 nucleotide sequences were generated to ascertain the VP7 genotype nature of the RUBV strains (Table 1). RUBV strains shared maximum VP7 identities (97·7–99·5% at nt level and 97·8–100% at aa level) with the novel bovine GBR Kolkata strains, and shared identities of 73·7–78·9% and 80·8–89·6%; 62·6–66·2% and 59·8–65·4%; 58·9–62·2% and 48·6–54·9% at nt level and aa level with other bovine, human and murine GBR. On phylogenetic analysis, RUBV strains clustered with Kolkata strains, and were distantly related to other bovine, human, and murine GBR (Fig. 3). Similarly, the partial NSP5 encoding gene sequences (variable lengths, ranging from 552 to 571 nt long) of RUBV strains exhibited high identities (93–99% at nt level and 97–100% at aa level) with the Kolkata strains, and shared low nt and aa identities of 48–62% and 48·9–50·6%; 62–64% and 48–49·1%; 77–78% and 81·9–83% with cognate sequences of human, murine and ovine GBR strains, corroborating the genetic relatedness between RUBV and the Kolkata strains.

Fig. 3. Phylogenetic analysis of the deduced VP7 amino-acid sequences (partial, amino acid 23–247) of representative RUBV strains with that of bovine, human and murine group B rotaviruses. The phylogenetic tree was constructed by the neighbour-joining method (random number generator seed of 111 and 1000 bootstrap trials). Bootstrap values have been mentioned. The Kolkata strains are DB101, DB176 and DB180. The tree was rooted with cognate sequence of atypical human rotavirus strain J19. Abbreviations: C-B, Chhapra-Bihar; P-B, Purnia-Bihar; D-J, Dumka-Jharkhand; G-U, Gorakhpur-Uttar Pradesh.
DISCUSSION
Contrary to GAR, the epidemiology of GBR in humans and animals remains largely unexplored [Reference Kapikian, Hoshino, Chanock, Knipe, Howley, Griffin, Lamb, Martin, Roizman and Straus1–Reference Sen, Raghunath and Nayak3]. Since the Chinese epidemics of 1982–1983, affecting >1 million of the population, human GBR have been primarily associated with sporadic cases of diarrhoea in adults, and rarely been detected in children [Reference Kobayashi, Naik, Ahmed and Kobayashi2]. Recently, our laboratory reported a shift in age preference of GBR infection from adults to children, revealing the importance of GBR as an aetiological agent of childhood diarrhoea [Reference Barman5]. In cattle, studies on GBR appear to be rare in calves compared to cows [Reference Saif, Jiang and Ramig20], and have been associated with sporadic cases of diarrhoea in calves from India [Reference Barman15], Japan [Reference Tsunemitsu8] and the United States [Reference Chang6, Reference Chinsangaram7]. In Japan, 11·5% (n=52) of calves were positive for GBR antibodies [Reference Tsunemitsu8], and from the United States, two separate surveillance studies reported infection rates of 5·6% (n=90) [Reference Chang6], and 81% (n=47) with repeated sampling from same animals [Reference Chinsangaram7]. In India, the three bovine GBR strains (Kolkata strains [Reference Barman15]) with a novel VP7 genotype were reported from a cattle market in Kolkata city during a surveillance study (1·5%, n=192) between November 2001 and August 2002. The present surveillance study (December 2003–March 2005) reports a significant increase in the prevalence of ‘Kolkata strain’-like novel GBR strains (RUBV strains, 13·0%, n=260) among calves with diarrhoea from the same Kolkata cattle market.
Calves sold in the market were traced to four districts in neighbouring states of West Bengal. In the past, the Kolkata GBR strains were detected from calves transported from Purnia and Chhapra districts, Bihar, Eastern India. Similarly, during the present study, 8, 11, 12 and 3 of the 34 GBR-infected calves were traced to Chhapra, Purnia, and Dumka districts of Bihar and Jharkhand in Eastern India, and Gorakhpur in Uttar Pradesh, Northern India. Since the samples for GBR were collected immediately after the arrival of the calves in the cattle market, the possibility of the calves acquiring the infection from the cattle market appeared unlikely. Similarly, as the overnight journey across the states to the cattle market was non-stop, acquisition of infection en route was also ruled out. GBR have an incubation period of 1–3 days, and calves begin to shed virus between 1 and 3 days after infection [Reference Chang6, Reference Tsunemitsu11]. On the other hand, the total time taken for the overnight journey to the cattle market ranged between 3 and 11 h (depending upon the distance of the district and state from Kolkata city). Since samples were collected immediately after the arrival of the calves in the cattle market, it is unlikely that the samples positive for GBR were a result of acquisition of infection during transit. Therefore, it might be possible that the calves had acquired the infection prior to their transportation to Kolkata, West Bengal. This was further supported by the presence of diarrhoea in 29 of the 34 calves (as revealed by case histories), positive for GBR prior to transportation to Kolkata. Calves arriving in the cattle market were being sold to farms located in various districts of West Bengal, and therefore, the possibility of transmission of these strains across different parts of West Bengal is always present. Earlier surveillance studies (1999, n=167 [Reference Samanta and Sarkar21]; 2000, n=112 [Reference Panja22]; 2002, n=140, and 2003, n=120 [data generated in our laboratory]) conducted in various districts of West Bengal revealed absence of bovine GBR. Interestingly, a recent surveillance study (2005, n=46) in a cattle shed (shelter for stray and debilitated cattle, 15 km from the cattle market) reported an identical GBR strain (LB51, accession no. DQ538516) from a calf (aged 4 months) with diarrhoea, raising concern over the probable spread of bovine GBR across West Bengal. However, since the GBR-infected calf strayed to the shed, we could not trace back to the origin of the strain.
GAR strains exhibiting VP7 aa identities ⩾90% belong to same genotype, and within a species, different genotypes have been reported [Reference Kapikian, Hoshino, Chanock, Knipe, Howley, Griffin, Lamb, Martin, Roizman and Straus1]. Applying the same classification system, bovine, human, and murine GBR strains qualified as separate genotypes, and unlike GAR, genotype specificity collated with species specificity. On the other hand, the Kolkata and RUBV strains exhibited low aa identities with other bovine GBR, and pointed towards existence of separate genotypes within a species for GBR. The genetic relatedness between the RUBV and Kolkata strains were further supported by sequence analysis of the NSP5 gene.
In conclusion, the present study provides vital insights into interstate transmission of bovine GBR infection within a country, and points towards the rapid increase in prevalence of novel GBR strains associated with disease condition in calves. In GAR, continuous surveillance studies encompassing different populations, and under various geographical settings have revealed an increase in prevalence of rare and/or novel VP7 genotypes, often in proportions (as in G9 and G12 strains), necessitating its inclusion in rotavirus vaccines [Reference Santos and Hoshino23]. A similar situation might be prevailing for GBR, and therefore, future surveillance studies should consider detection and molecular characterization of GBR along with GAR in humans and animals.
ACKNOWLEDGEMENTS
The research was financially supported by Indian Council of Medical Research (ICMR), Government of India. S. Ghosh and V. Varghese were supported by Senior Research Fellowships from the Indian Council of Medical Research, Government of India.
DECLARATION OF INTEREST
None.









