Introduction
Norovirus and rotavirus are the most common causative agents of pediatric acute gastroenteritis worldwide, including in South Korea. The rotavirus infection rate in children has decreased since the introduction of a vaccine in 2007–2008 [Reference Jin1]. Norovirus-related gastroenteritis has an important impact on public health worldwide. However, official public health reports likely largely underestimate the actual incidence. Therefore, public health surveillance and thorough statistical data analysis are needed [Reference LeBlanc2].
Norovirus infection is accompanied by vomiting and diarrhoea after a latency period of 12–48 h and the virus is transmitted person to person via the fecal-oral route or via a contaminated environment [Reference Fretz3]. Norovirus outbreaks have been reported frequently in closed environments, such as facilities, schools, hospitals and cruise ships and often originate from contaminated groundwater [Reference Cho4]. The incidence of norovirus among children is very high; norovirus causes 200 000 deaths in children younger than 5 years of age annually in developing countries [Reference Pothier and Kaiser5]. The elderly and immunocompromised patients are also vulnerable to norovirus and are at risk of dying from infection [Reference Pothier and Kaiser5].
According to the Centers for Disease Control and Prevention, there are 21 million norovirus cases in the USA annually, accounting for 60% of patients with acute gastroenteritis [Reference Robilotti, Deresinski and Pinsky6]. In the USA alone, each year, 570–800 people die, 56 000–71 000 people are hospitalised and the total hospitalisation cost is about 500 million. Medical costs and lost productivity are estimated at 2 billion [Reference Lee7]. According to the Foodborne Viruses in Europe report, 7637 norovirus outbreaks occurred in 13 European countries from July 2001 to June 2006, mostly in Germany and the UK, with 3808 (49.9%) and 1938 (25.4%) outbreaks, respectively [Reference Kroneman8].
Norovirus was reported in South Korea for the first time in 1999 and the National Institute of Health has been monitoring acute diarrhea diseases since 2000 [Reference Gong9]. In 2006, norovirus caused 2400 infection cases at 31 school cafeterias throughout the country [Reference Koo10]. In 2008, approximately 130 infection cases in an elementary school in Incheon were reported [Reference Yu11]. According to the Korea Food and Drug Administration, the number of norovirus outbreaks and patients in Korea has increased significantly since 2000 [Reference Cheon12].
The development of a vaccine or therapy for norovirus is hampered by the lack of a suitable cell culture system and animal models [Reference Duizer13]. Previous studies on norovirus have mainly relied on electron microscopy, antibody investigation and reverse transcription-polymerase chain reaction (RT-PCR) [Reference Koo10].
Norovirus belongs to the family Caliciviridae [Reference Patel14]. It has a single-stranded, positive-sense RNA genome of approximately 7.5 kb [Reference Vega15]. The genome comprises three open reading frames (ORF). ORF1 encodes non-structural proteins, such as NTpase, protease and RNA-dependent RNA polymerase (RdRp). ORF2 encodes major capsid protein (VP1). ORF3 encodes minor capsid protein (VP2) [Reference Kim16]. Noroviruses are divided into seven genogroups (GI–GVII) and are classified into more than 40 genotypes [Reference Vinje17]. GI, GII, GIV can infect humans. GII is the most common genogroup [Reference White18] and GII.4 is the most predominant genotype, with new variants occurring in 2–3-year cycles. GII.4 viruses have a larger susceptible population than viruses from other genotypes as they can bind a wider range of histo-blood group antigens, which have been suggested to be attachment factors [Reference Eden19]. The high rate of evolution of GII.4 viruses leads to the emergence of new antigenic variants [Reference Lindesmith, Donaldson and Baric20].
GII.17 occurred at a low frequency for many years [Reference Han21]. Recently, a GII.17 norovirus variant emerged [Reference Pogan, Dulfer and Uetrecht22] and globally became more dominant than GII.4, which had been dominant in China and Hong Kong for more than 20 years. The novel variant has been sporadically reported in Italy and the USA [Reference Chan23]. In Japan in 2014, strains with an RdRp sequence different from that of existing GII.17 were found. This variant with newly identified RdRp was assigned GII.P17-GII.17 by the web-based norovirus typing tool NoroNet [Reference Matsushima24]. In this study, we sequenced the whole genome of GII.P17-GII.17 strains discovered in 2013 and we confirmed amino acid mutations through various comparative analyses with other global isolates.
Materials and methods
Ethics statement
Stool samples of patients with acute gastroenteritis symptoms were provided by Waterborne Virus Bank. Due to difficulties in tracing the exact records of the young patients at the donation hospital, informed consent could not be obtained from the patients' parents. The institutional review board of Songeui Medical Campus, The Catholic University of Korea (MC18SESI0063) reviewed and approved the use of the samples for research purposes as this study had no impact on the patients. All experimental work and sample collection were supervised by the institutional review board.
Stool sample preparation
In total, 204 stool samples were collected from children of 0–13 years of age with fever and diarrhea between January 2013 and December 2013. Frozen stool samples were diluted with 10% phosphate-buffered saline and centrifuged at 13 000 × g for 10 min at 4 °C. The supernatant was separated and stored at −80 °C until analysis.
Viral RNA extraction
Viral RNA was extracted from 140 µl of stool sample using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The RNA was stored at −80 °C until use in RT-PCR.
RT-PCR analysis
Norovirus-positive specimens were detected by RT-PCR using the GI-F1M–GI-R1M and GII-F1M–GII-R1M primer sets [Reference Lee7] (Table 1) and a One-Step RT-PCR kit (Qiagen, Hilden, Germany). Thermal cycles were as follows: 50 °C for 30 min, 95 °C for 15 min, followed by 40 cycles of 94 °C for 60 s, 55 °C for 60 s, and 72 °C for 60 s and a final step at 72 °C for 10 min. To analyse the whole-genome sequence of GII.17 norovirus, 14 pairs of primers were newly designed (Table 2). Thermal cycles were the same as above. PCR products were analysed by electrophoresis in ethidium bromide-stained 2% agarose gels.
Table 1. Sequence information for primers used for RT-PCR assays

Table 2. Newly designed primers used

The primers were based on the kawasaki323 strain (AB983218), CAU-55 strain (KU561250).
GSP1, GSP2 and Nested GSP were based on the CMC-01 strain.
Determination of the 5′- and 3′-ends of norovirus genomic RNA
To determine the 5′-ends of norovirus genomic RNA, rapid amplification of cDNA ends (RACE) was performed using the 5′ RACE System for Rapid Amplification of cDNA Ends Version 2.0 Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's recommendations. Three primers (GSP1, GSP2 and nested GSP) were designed based on ORF1 of CMC-1 for RACE (Table 2). To obtain the exact sequence of the 3′-end of the genomic RNA, cDNA was synthesised by reverse transcription using 3′-oligo (dT)-anchor-R (Table 2). The second PCR was conducted using the ORF3_2F and 3′-anchor-R primers (Table 2) under the following conditions: 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min, followed by 72 °C for 7 min.
Cloning and sequencing of the complete genome
GII.17 norovirus PCR product was extracted from a 2% agarose gel using the HiYield Gel/PCR DNA Fragments Extraction Kit (Real Biotech, Taipei, Taiwan) and was cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). The cloned vectors were transformed into Escherichia coli DH5α competent cells (RBC Bioscience) according to the manufacturer's instructions and selected at 37 °C for 16–18 h on Luria–Bertani agar plates (Duchefa, Haarlem, the Netherlands) containing 40 mg/ml X-gal, 0.1 mM isopropyl-β-d-thiogalactoside and 50 mg/ml ampicillin. Selected clones were inoculated into LB broth (Duchefa) and cultured overnight at 37 °C under shaking at 200 rpm. Plasmid DNA was purified using a HiYield Plasmid Mini Kit (RBC Bioscience) and sequenced (Macrogen, Seoul, South Korea). The sequencing results were analysed using the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information (NCBI); National Institutes of Health, Bethesda, MD, USA).
Phylogenetic analysis
Nucleotide sequences of GII.17 were aligned and classified by ORF1, ORF2 and ORF3. Phylogenetic analysis of the GII.17 genome sequences was performed using sequences of norovirus strains published in the NCBI database, using Molecular Evolutionary Genetic Analysis software (MEGA version 7.0).
Results
Norovirus detection and whole genome sequencing
As a result of screening 204 stool samples, 11 samples (5.39%) were determined to contain norovirus belonging to genogroup GI and 60 samples (29.41%) were found to contain norovirus belonging to GII. In GI samples, we detected genotypes GI.4 (n = 7; 3.43%) and GI.5 (n = 4; 1.96%). In GII samples, we identified GII.4 (n = 45; 22.06%), GII.6 (n = 12; 5.88%) and GII.17 (n = 3; 1.47%). The three GII.17 viral specimens were named CMC-01, CMC-02 and CMC-03. All three viruses were subjected to whole genome sequencing. The genome of GII.17 has a total length of 7512 nt and consists of ORF 1 (5109 nt), ORF 2 (1623 nt) and ORF 3 (780 nt). The genome sequences obtained in this study have been deposited in NCBI (MK282256, MK282257 and MK282258).
Phylogenetic analysis
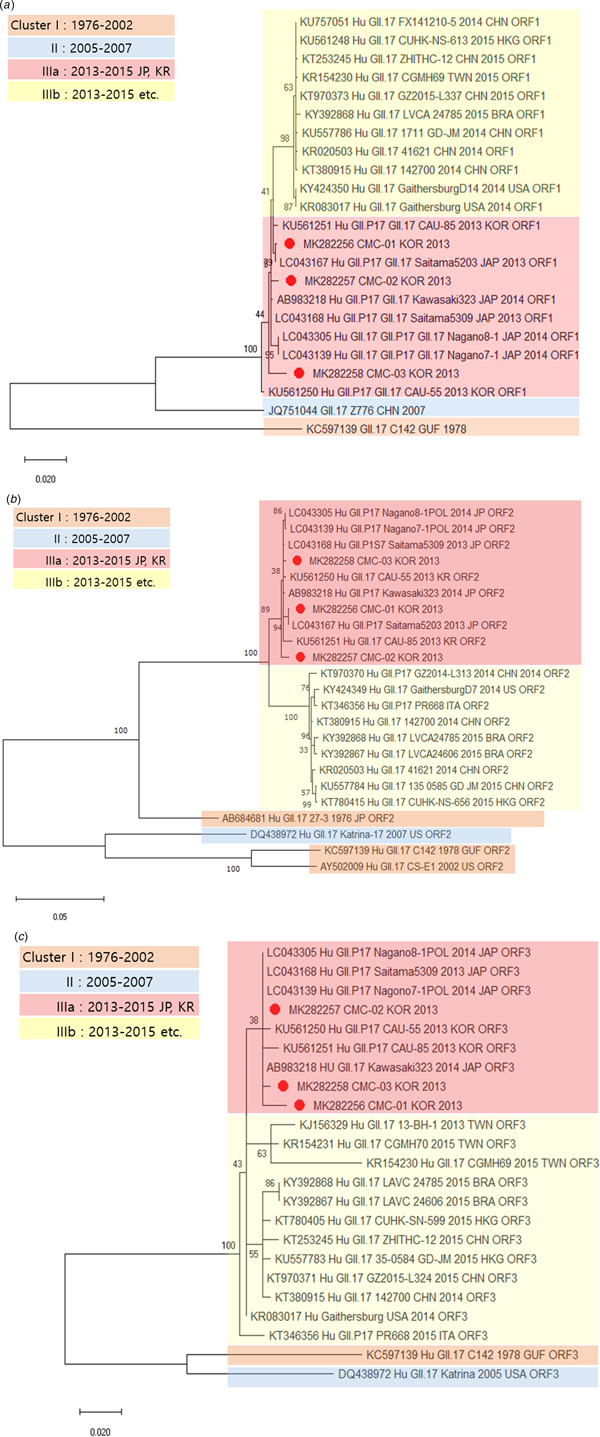
Genetic relationships between the viral specimens from this study and norovirus strains reported in GenBank were determined on the basis of phylogenetic analysis. The whole genome sequences of CMC-01, CMC-02 and CMC-03 were classified as norovirus GII.17 and showed maximum identity (99.5%) with the strain Saitama5203 (LC043167) (Fig. 1). The sequences of CMC-01, CMC-02, CMC-03 were compared and analysed with the sequences of isolates reported in various countries, including Brazil, Japan and China, from 1976 to 2016. Prior to the analysis, we divided cluster I (1976–2002), cluster II (2005–2007), cluster IIIa (2013–2015) and cluster IIIb (2013–2015) by year and country. IIIa strains are found in Korea and Japan, IIIb strains are found in countries other than Korea and Japan, and CMC-01, CMC-02 and CMC-03 were classified in cluster IIIa. Next, we constructed phylogenetic trees based on amino acid sequences. The ORF1 sequence of CMC-01 showed 99.5% identity with that of Saitama5203 (LC043167). The ORF1 sequences of CMC-02 and CMC-03 showed 99.5% and 99.7% identity with that of Saitama5309 (LC043168), respectively (Fig. 2a). For ORF2, CMC-01 showed the highest identity of 99.9% with Saitama5203 and CMC-02 and CMC-03 showed 99.3% and 99.8% identity with Saitama5309 (Fig. 2b). ORF3 of CMC-01 (99.3%), CMC-02 (99.7%) and CMC-03 (99.7%) showed the highest similarity with that of Saitama5309 (LC043168), Kawasaki323 (AB983218) (Fig. 2c). A phylogenetic tree was constructed with strains collected from 1976 to 2015. ORF1, ORF2 and ORF 3 of the three detected in this study all fell in cluster IIIa and were further classified as GII.P17–GII.17, which is a variant of GII.17. All three viruses were very similar to Saitama5309 (LC043168), Kawasaki323 (AB983218), Nagano8-1 (LC043305) and Nagano7-1 (LC043139).

Fig. 1. Phylogenetic analysis of the full-length sequences of the GII type detected determined by genotyping and reference strains isolated worldwide.

Fig. 2. Phylogenetic analysis of norovirus based on nucleotide sequences. The trees were constructed with the neighbour-joining method. Phylogenetic trees based on (a) amino acid sequence of ORF1, (b) amino acid sequence of ORF2 and (c) amino acid sequence of ORF3. CMC-01, CMC-02 and CMC-03 are indicated by red circles.
VP1 sequence alignment
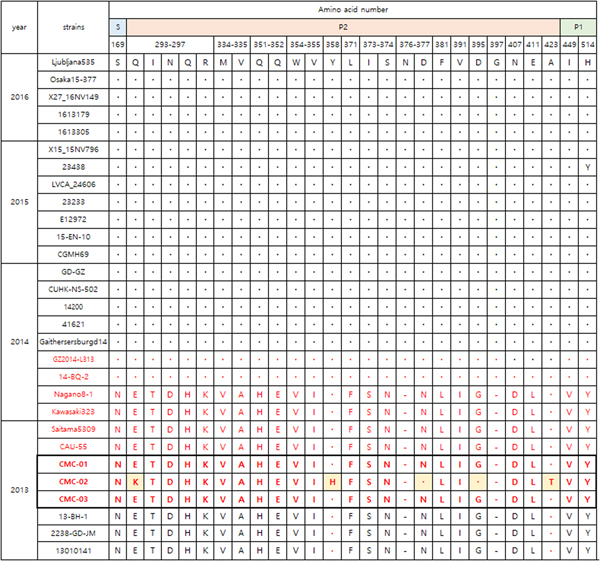
Figure 3 shows the VP1 amino acid substitution pattern in our viral specimens compared with 12 strains collected from 1976 to 2015 based on alignment. CMC-01, CMC-02 and CMC-03 were classified with GII.P17-GII.17 strains collected in 2013–2014. Specifically, the GII.17 strains 14-BQ-2 and GZ2014-L313 discovered in 2014, had 28 amino acid alterations in the P2 domain (Fig. 4). CMC-01 and CMC-03 were similar to GII.P17-GII.17 strains and CMC-02 had five amino acid substitutions (Glu293Lys, Tyr360Lys, Asn379Asp, Gly397Asp and Ala425Thr). Comparative analysis of the 2013 and 2014 GII.17 strains revealed 21 substitutions, especially in the P2 domain (Glu293Gln, Thr294Ile, Asp295Asn, Gln298His, Asp301Asn, Val336Met, Ala337Val, His353Gln, Glu354Gln, Val356Trp, Ile357Val, Phe373Leu, −378Asn, Ser375Ile, Asn376Ser, Asn379Asp, Leu383Phe, Val393Ile, Gly397Asp, Asp409Asn and Leu413Glu). When comparing GII.P17-GII.17 and GII.17 strains, the substitutions Gln298His, −378Asn and Val393Ile were specific to GII.P17-GII.17. In addition, substitutions including Asn169Ser in the shell (S) domain and Asp410Asn, Leu414Glu and Tyr516His in the P1 domain were detected.

Fig. 3. Amino acid substitutions in the viruses in this study (GII.17; black colour, GII.P17-GII.17; red colour). Alignment of VP1 amino acid sequences of CMC-01, CMC-02 and CMC-03 strains. Dots indicate sequence identity among sequences presented. Dashes indicate deletions/insertions of amino acid residues. LVCA24606 (KY392867), CGMH69 (KR154230), GaithersburgD14 (KY424350), 41621 (KR020503), Nagano8-1 (LC043305), Kawasaki323 (AB983218), CAU-55(KU561250), Saitama5309 (LC043168), 13010141 (KU757046), 2238 (KU557788), Katrina-17 (DQ438972), 27-3 (AB684681).

Fig. 4. Comparison of major amino acid substitutions in viral specimens of 2013–2016 (GII.17; black colour, GII.P17-GII.17; red colour). Dots indicate sequence identity among sequences presented. Dashes indicate deletions/insertions of the amino acid residues. Amino acid numbering is based on the sequence of the Saitama5309 strain. Ljubljana535(KX134671), Osaka15-377(LC148852), X27-16NV149 (KX371112), 1613179(KU953395), 1613305(KU953397), X15_15NV796 (KX371109), 23438 (KX216804), LVCA_24606 (KY392867), 23233 (KX216793), E12972 (KU587628), 15-EN-10 (KT732275), CGMH69 (KR154230), GD-GZ (KU557801), CUHK-NS-502 (KT780399), 14200 (KT380915), 41621 (KR020503), GaithersburgD14 (KY424350), GZ2014-L313 (KT970370), 14-BQ-2 (KT906670), Nagano8-1 (LC043305), Kawasaki323 (AB983218), Saitama5309 (LC043168), CAU-55 (KU561250), 13-BH-1 (KJ156329), 2238-GD-JM(KU557788), 13010141 (KU757046).
Discussion
Norovirus globally is one of the important causal agents of acute gastroenteritis. Although norovirus causes economical and public-health problems, a thorough study of norovirus is hampered by the lack of cell culture systems or animal models. In infants and the elderly, illness can last longer [Reference Lee and Pang25]. GII.4 accounts for 70–80% of norovirus gastroenteritis since the mid-1990s [Reference Medici26]. Since 1996, GII.4 variants have periodically appeared every 2–3 years [Reference Eden19] and GII.4 is the major genotype circulating in the community and medical environment [Reference Chan27]. A GII.P17-GII.17 norovirus strain was discovered in Japan during the winter season of 2014–2015.
In this study, we screened 204 stool samples collected from children with symptoms of acute gastroenteritis between January 2013 and December 2013 for the presence of norovirus. The collected strains were classified as GII.17 by BLAST and were identified as GII.P17-GII.17 according to the sequences of viral RdRp. Other GII.P17-GII.17 strains include Saitama 5309 (LC043168) and Kawasaki 323 (AB983218). Sequence analysis of ORF1–3 revealed a very high similarity between our viruses and previously collected GII.P17-GII.17 strains. CMC-01 is very similar to Saitama5203 (LC043167) and CMC-02 and CMC-03 showed high sequence identity with Saitama5309 (LC043168). The GII.17 strain detected in South Korea was phylogenetically related with strains detected in the USA, Italy, China, Japan and Hong Kong. Interestingly, the GII.3 strain discovered in the 2000s has an evolutionary relationship with the recently discovered GII.17 strain [Reference Dang Thanh28]. Norovirus VP1 consists of an S domain that forms a scaffold enveloping the viral RNA and a P domain composed of subdomains P1 and P2 [Reference Beier29]. The S domain has the most stable conserved sequence. The P1 domain is less variable than P2 domain and the P2 domain is the most exposed and the most variable in structure [Reference Prasad30]. Therefore, among highly similar strains, variations in the P2 domain sequence gives rise to different P2 domain structures. CMC-01 and CMC-03 showed very close similarity to previously collected strains, whereas CMC-02 showed six amino acid substitutions (Figure 3; Glu293Lys, Tyr360His, Asn379Asp, Gly397Asp, Ala425Thr and Glu473Asp). Figure 4 shows amino acid substitutions in the VP1 region of viruses collected in 2013–2016. GII.17 viruses collected after 2014 showed amino acid sequence variants different from those occurring before 2014 (Glu293Gln, Thr294Ile, Asp295Asn, His296Gln, Lys297Arg, Val334Met, Ala335Val, His351Gln, Glu352Gln, Val354Trp, Ile355Val, Phe371Leu, Asn373Ile, Asn374Ser, − 376Asn, Asn377Asp, Leu381Phe, Ile391Val, Gly395Asp, − 397Gly, Asp407Asn, Leu411Glu). The hypervariable P2 domains of VP1 of the CMC strains were different from those of GII.17 viruses reported after 2014, but similar to those of GII.P17-GII.17 and GII.17 viruses collected before 2014. Even though strains 14-BQ-2 (KT906670) and GZ2014-L313 (KT970370) were classified as GII.P17-GII.17, their P2 domain is similar to that of GII.17 viruses collected after 2014. Thus, it can be assumed that the timing of appearance of 14-BQ-2 (KT906670) and GZ2014-L313 (KT970370) may be similar to that of GII.17, based on P2 domain similarity. Thus, GII.P17-GII.17 was prevalent in 2013–2014 and it is likely that the P2 domain changed at that time. A GII.P17-GII.17 norovirus strain was discovered in Japan during the winter season of 2014–2015. Since then, it has been detected in Asian countries at high frequency and in other continents at low frequency [Reference Medici26]. However, GII.P17-GII.17 strains (KU561250, KU561251) were collected in Korea in 2013; thus, this type may have been prevalent in Korea before it occurred in Japan. This sequence data will be useful for comparisons with full-length norovirus sequences of other strains identified worldwide. Moreover, the information acquired from the whole-genome sequences of strains CMC-01 and CMC-03 may prove useful for advancing basic research toward the elucidation of gene functions, the prediction of newly appearing pandemic variants via comparison with norovirus in neighbouring countries and in vaccine development. Overall, broadening the information and genetic resources of noroviruses circulating globally will have important benefits for public health and help to identify new emerging strains of norovirus.
Author ORCIDs
S. Y. Paik, 0000-0003-2748-6699.
Acknowledgement
This work was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (grant number HI15C1781); and the Korea Ministry of Environment (MOE) as a Public Technology Program based on Environmental Policy (grant number 2016000210002).
Conflict of interest
None.