Lactic acid bacteria comprise a heterogeneous group of micro-organisms that produce lactic acid as a major end product of carbohydrate fermentation. Lactic acid bacterial strains are historically important because of their utility in food preservation and fermentation. Some strains are used as probiotics defined as live micro-organisms which, when administered in adequate amounts, confer a health benefit on the host(Reference Fuller1, 2). Probiotic effects include modulation of the innate and adaptive immune systems, suppression of intestinal infections and alleviation of food allergies(Reference Delcenserie, Martel and Lamoureux3, Reference Leblanc, Fliss and Matar4). Since some probiotics enhance cell-mediated cytotoxic activity against tumour cells and virus-infected cells(Reference Takeda and Okumura5, Reference Namba, Hatano and Yaeshima6), it is possible that they could help protect consumers from the risk of tumour development and infectious diseases.
Lactobacillus brevis KB290 (KB290), which was isolated from the traditional Japanese pickle Suguki, is safe for humans, tolerates gastrointestinal juices and improves gut health(Reference Nobuta, Inoue and Suzuki7), as well as being useful for an early intervention in the irritable bowel syndrome(Reference Murakami, Habukawa and Nobuta8) – characteristics that meet the criteria for a probiotic strain. Humans who ingest KB290 show enhancement of interferon-α production capacity(Reference Kishi, Uno and Matsubara9). Oral administration of 1 × 109 colony-forming units (cfu)/d of KB290 for 14 d enhanced cell-mediated cytotoxic activity in mouse splenocytes (Y Fukui and K Katsura, unpublished results).
Natural killer (NK) cells act as cytolytic effector cells of the innate immune system. Activation of NK cells is induced by direct recognition of cells infected with viruses via NK receptors as well as by cytokines produced by innate immune cells, such as dendritic cells (DC)(Reference Fernandez, Lozier and Flament10, Reference Lucas, Schachterle and Oberle11). CD8+ cytotoxic T cells can also recognise and eradicate virally infected cells and tumours. DC and macrophages elicit CD8+ cytotoxic T-cell activation by the interaction between MHC class I and the T-cell antigen receptor(Reference Edidin12). Lactic acid bacterial strains initiate NK–DC interactions via DC maturation and, as a consequence, NK cells increase their cytolytic potential(Reference Rizzello, Bonaccorsi and Dongarra13). However, the mechanism by which lactic acid bacteria modulate the function of effector cells in vivo is not fully understood because of a complex network of immune cells.
DNA microarray technology allows us to evaluate the expression of many genes at once and how they are up- and down-regulated in a particular type of cell or tissue. Nutrigenomics(Reference Kato and Kimura14), an application of genomic technology to nutrition research, is also becoming important to the food industry, and many nutrigenomic studies have utilised new approaches such as bioinformatics to understand how nutrients influence gene expression. This technology has been used to study how probiotics modulate the immune system in vivo focused on intestinal mucosal immunity or the anti-inflammatory effect(Reference Nerstedt, Nilsson and Ohlson15, Reference Reiff, Delday and Rucklidge16). Here, we use the technology to investigate immune responses to ingested KB290 by focusing on cell-mediated cytotoxicity.
Materials and methods
Bacterial strain
KB290 was deposited as strain L. brevis JCM 17,312 in the Japan Collection of Microorganisms and has been maintained at Research Institute, Kagome Company Limited (Tochigi, Japan). Recognised as safe for human consumption(Reference Nobuta, Inoue and Suzuki7), KB290 is used commercially in beverages and supplements.
Animal experiments
Specific pathogen-free female BALB/c mice, aged 9 weeks, were purchased from Charles River Laboratories Japan, Inc. They were housed at 20–24°C and 45–65 % humidity in an animal laboratory with a 12 h light–12 h dark cycle timed from 07.30 hours. They were divided into two groups (n 6) with equal mean body weights and were fed a commercial normal diet (CE-2; CLEA Japan, Inc.) and sterile water during a 1-week acclimatisation period. Thereafter, lyophilised KB290 was added to the treatment group diet (3 × 109 cfu/g), while 5·8 % (w/w) potato starch (Nippon Starch Chemical) was added to the control group diet. Daily intake of the KB290-containing diet per treated mouse was 3·63 g, or about 1 × 1010 cfu, as the combination of a high dose and the short duration of the treatment showed cell-mediated cytotoxicity reproducibly (Y Fukui and E Sasaki, unpublished results).
On day 1, mice were euthanised, their spleens were sampled and cell-mediated cytotoxic activity was measured in splenocytes before analysing gene expression using DNA microarrays.
The Animal Care and Use Committee of the Institute of Kagome Company Limited approved all protocols, which were in accordance with the guidelines established by the Japanese Society of Nutrition and Food Science (Law and Notification 6 of the Japanese Government).
YAC-1 target cells
Mouse lymphoma YAC-1 cells (Japanese Collection of Research Bioresources) were maintained in Roswell Park Memorial Institute-1640 (RPMI) medium (Sigma) supplemented with 10 % (v/v) fetal bovine serum (Nippon Biotest), 5 mm-HEPES (Sigma), 1 % penicillin–streptomycin solution (p4458; Sigma) and 100 μg streptomycin/ml (Sigma) (complete RPMI-1640 medium). NK cells(Reference Savary and Lotzova17), CD8+ cytotoxic T cells(Reference Stewart and Hoskin18) and macrophages(Reference Ribeiro-Dias, Russo and Marzagao Barbuto19) showed cytotoxicity against the cells.
Preparation of splenocytes
Individual mouse spleens were placed into 5 ml minimum essential medium Eagle (Sigma) containing 1 % penicillin–streptomycin solution (p4458; Sigma), and crushed with frosted slide glass (Matsunami Trading) to obtain single-cell suspensions. The cells were passed through a sterile 70 μm nylon mesh filter (Becton Dickinson) and transferred to a 15 ml tube (Becton Dickinson) filled with complete RPMI-1640. They were centrifuged at 430 g for 3 min, collected, washed once in PBS (Invitrogen), counted with an automatic blood cell counter (Nihon Kohden) and adjusted the final concentration to 4 × 106 cells/ml in complete RPMI-1640 medium.
Cell-mediated cytotoxicity assay
Flow cytometry measured the cell-mediated cytotoxic activity of splenocytes to calculate the percentage of the killed cells stained with propidium iodide (PI; Sigma)(Reference Johann, Blumel and Lipp20). YAC-1 cells (106 cells/ml) were labelled by incubation for 10 min with 20 μg/ml of 3,3′-dioctadecyloxacarbocyanine perchlorate (Dio; Sigma) at 37°C in a 5 % CO2 atmosphere. After washing them three times with PBS, the cells (105 cells/ml) were resuspended in complete RPMI-1640 medium and mixed 50 or 100 μl of the splenocyte suspension (4·0 × 106 cells/ml) with 100 μl of the YAC-1 cell suspension to achieve effector:target cell ratios of 20:1 and 40:1, and then added PI (25 μg/ml). To enhance cell contact, the samples were centrifuged for 5 min at 430 g and incubated for 2 h at 37°C in 5 % CO2. Target cell lysis was determined using a flow cytometer (FACSCalibur™; Becton Dickinson) and analysed with CELLQuest software (Becton Dickinson). The percentage of the dead target cells was calculated (%T d) as (% Dio+ PI+ cells/% Dio+ cells) × 100 and cell-mediated cytotoxic activity was estimated as %T d (cultured with the effector cells) − %T d (cultured without the effector cells). Each sample was assayed in triplicate and the means were recorded.
DNA microarray assay
Total RNA was isolated from each spleen sample with TRIzol reagent (Invitrogen), according to the manufacturer's instructions, and purified using the RNeasy Mini Kit (Qiagen) and checked the quality and quantity by agarose gel electrophoresis and spectrophotometry, respectively. Total RNA from individual samples was subjected to DNA microarray analysis as described previously(Reference Suyama, Okada and Ishijima21). Briefly, 100 ng of the purified total RNA were used to synthesise complementary DNA, and then biotinylated amplified RNA (aRNA) was transcribed with T7 RNA polymerase using the GeneChip 3' IVT Express Kit (Affymetrix). Agarose gel electrophoresis assayed aRNA quality, ascertaining its suitability for the experiments. aRNA was fragmented and then hybridised to an Affymetrix GeneChip mouse genome 430 2.0 array (Affymetrix) to determine the expression of over 45 000 probe sets fully covering the mouse genome, in accordance with the manufacturer's instructions. The array was hybridised at 45°C for 16 h, washed and stained with phycoerythrin. Fluorescence signals were scanned with the Affymetrix GeneChip System and Affymetrix GeneChip Command Console software was used to reduce the images to the intensity values for each probe (CEL files). All microarray data were submitted to the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; GEO Series ID GSE41127).
Quantitative RT-PCR
Single-strand complementary DNA was synthesised from 20 ng of total RNA using the PrimeScript RT reagent kit (Takara Bio). Quantitative RT-PCR analysis was performed on the 7000 Real-Time PCR System (Applied Biosystems) using SYBR Premix Ex Taq (Perfect Real Time; Takara Bio), according to the manufacturer's instructions. The reaction conditions were as follows: initial denaturation at 95°C for 30 s followed by forty cycles of denaturation at 95°C for 5 s and 60°C for 31 s. The primer sequences were as follows: IL-18 (Il18), forward 5′-AAGACTCTTGCGTCAACTTCAAGGA-3′ and reverse 5′-AGTCGGCCAAAGTTGTCTGATTC-3′; DNAX activation protein of 12 kDa (Dap12), forward 5′-CCGGAAACAACACATTGCTGAG-3′ and reverse 5′-GCCTCTGTGTGTTGAGGTCACTGTA-3′; lymphocyte-specific protein tyrosine kinase (Lck), forward 5′-TGGGACCTTCACCATCAAGTCA-3′ and reverse 5′-GTCAGGTCTCACCATGCGGTAG -3′; purine-nucleoside phosphorylase 1 (Pnp), forward 5′-CCAACTTTGAGACTGTGGCAGA-3′ and reverse 5′-CATGACAACCTTGTTCGTAATGAG-3′; glyceraldehyde-3-phosphate dehydrogenase (Gapdh), forward 5′-AAATGGTGAAGGTCGGTGTG-3′ and reverse 5′-TGAAGGGGTCGTTGATGG-3′. Fold induction values were calculated using the 2− ΔΔCt method and target gene expression was normalised to Gapdh.
Flow cytometric analysis
Flow cytometry determined the proportion of marginal zone macrophages (MZM) in the spleen. Incubating for 1 min at room temperature in 1 ml Red Blood Cell Lysing Buffer (Sigma) lysed erythrocytes from splenocyte suspension (106 cells/ml) before wash in ice-cold PBS ( − ) supplemented with 1 % (w/v) bovine serum albumin (Wako) and 0·1 % (w/v) sodium azide (staining buffer; Wako). The cells were treated with Rat Anti-Mouse CD16/CD32 (Clone 2.4G2; Becton Dickinson GmbH) for 5 min on ice to block the Fc receptor and then stained the cells with R-phycoerythrin (PE)-conjugated rat anti-mouse macrophage receptor with collagenous structure (MARCO, Clone ED31; AbD Serotec), Alexa Fluor® 647-conjugated rat anti-mouse CD209b antigen (SIGNR1, Clone ER-TR9; AbD Serotec) and fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse H-2 (Clone M1/42; BioLegend) on ice for 20 min. After washing the cells using staining buffer, the samples were analysed using the flow cytometer with CELLQuest software (Becton Dickinson).
Data analysis
Student's t test was applied to compare cell-mediated cytotoxic activity, quantitative RT-PCR and the proportion of MZM of the experimental v. the control group using SPSS 15.0 for Windows (SPSS Japan, Inc.).
The CEL files were quantified with the Factor Analysis for Robust Microarray Summarization (FARMS) algorithm(Reference Hochreiter, Clevert and Obermayer22) using statistical language R (version 2.12.1) and Bioconductor 2.7(Reference Gentleman, Carey and Bates23). To detect the differentially expressed genes (DEG) between the control group and each of the KB290 groups, the rank products(Reference Breitling, Armengaud and Amtmann24) method was used as a non-parametric statistic(Reference Kadota, Nakai and Shimizu25). To identify functional classes of the DEG according to Biological Process in Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, enrichment analyses were performed using the Expression Analysis Systematic Explorer (EASE) score, the modified Fisher's exact test P value(Reference Hosack, Dennis and Sherman26) and Benjamini & Hochberg(Reference Benjamini and Hochberg27) false discovery rate corrections using the Database for Annotation, Visualization, and Integrated Discovery(Reference Huang da, Sherman and Lempicki28), a Web-accessible program, in accordance with the manuals available at the web site (http://david.abcc.ncifcrf.gov/home.jsp).
Results
Cell-mediated cytotoxicity
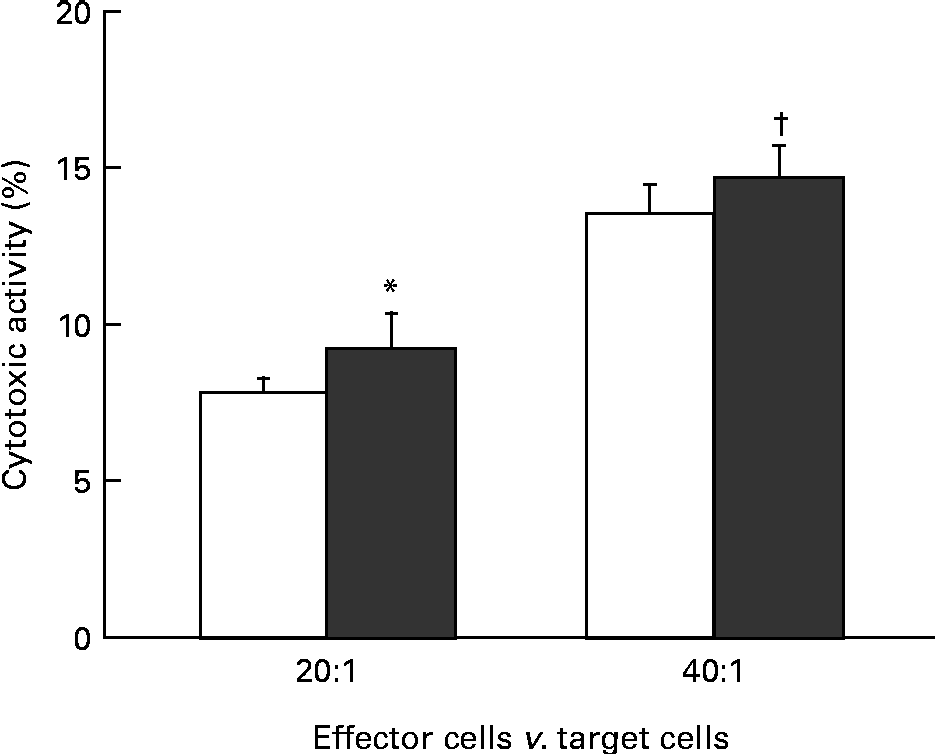
The effect of KB290 feeding for 1 d on cell-mediated cytotoxicity was investigated. Fig. 1 shows the cell-mediated cytotoxic activity of splenocytes from mice fed the KB290 or control diet. The activity was significantly (P <0·05) higher in the treated group at an effector:target cell ratio of 20:1, while the activity was marginal (P =0·067) at an effector:target cell ratio of 40:1.

Fig. 1 Cell-mediated cytotoxicity associated with KB290 feeding. Splenocytes were collected 1 d after the commencement of feeding a diet containing Lactobacillus brevis KB290 (KB290) (3 × 109 colony-forming units/g; ■) or potato starch (□) and used as effector cells against YAC-1 target cells (1 × 104 cells/well). The effector:target cell ratios were 20:1 and 40:1. Values are means, with standard deviations represented by vertical bars (n 6). * Mean value was significantly different compared with the control group (P< 0·05; Student's t test). † Mean value was marginally different from that of the control group (P= 0·067).
Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes analysis of DNA microarrays
To elucidate the mechanisms by which KB290 enhances cell-mediated cytotoxic activity, we used DNA microarrays and investigated gene expression changes in the spleen of mice ingested KB290. The rank products method combined with a qFARMS preprocessing algorithm nominated 395 up-regulated and 450 down-regulated probe sets in the group fed KB290 for 1 d (false discovery rate < 0·05). Of those, twelve probe sets were eliminated that were assigned as having both increased and decreased expression (this can be a property of the rank products method, which returns rank products statistics with false discovery rates for both up- and down-regulated probe sets independently). Finally, 383 up-regulated probe sets (327 unique genes) and 438 down-regulated probe sets (347 unique genes) were extracted. Full lists of the DEG are shown in Tables S1 and S2 (available online).
The GO and KEGG analysis identified functional classes of the DEG. The up- and down-regulated DEG fell into significantly (false discovery rate-corrected P <0·05) enriched GO terms (135 for the up-regulated genes summarised into fifty-three GO terms by removing redundant GO terms (Fig. 2) and fifteen for the down-regulated genes (Fig. 3)) and into significantly enriched KEGG pathways (eight for the up-regulated genes and two for the down-regulated genes (Table 1)). The over-represented GO terms and KEGG pathways among the up-regulated genes were involved in many immunologically relevant terms and pathways.

Fig. 2 Significantly enriched Gene Ontology (GO) terms found in the up-regulated genes by Lactobacillus brevis KB290 (KB290) feeding (P< 0·05). * GO term with no P value means not significant. † False discovery rate (FDR)-corrected P values of the GO terms appearing in the deepest hierarchy.

Fig. 3 Significantly enriched Gene Ontology (GO) terms found in the down-regulated genes by Lactobacillus brevis KB290 (KB290) feeding (P< 0·05). * GO term with no P value means not significant. † False discovery rate (FDR)-corrected P values of the GO terms appearing in the deepest hierarchy.
Table 1 Significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway found in the differentially expressed genes (DEG) by Lactobacillus brevis KB290 (KB290) feeding (P<0·05)
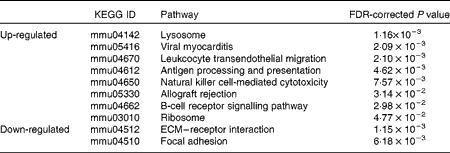
FDR, false discovery rate; ECM, extracellular matrix.
Gene expression profiles associated with cell-mediated cytotoxicity
GO terms appearing deeper in the hierarchical structure show more specificity, so they are more important. The deeper-level terms associated with cell-mediated cytotoxicity for each cluster of the hierarchy were searched, and the positive regulation of T-cell-mediated cytotoxicity and the antigen processing and presentation of peptide antigen via MHC class I in the immune system process were found. Almost all the up-regulated genes involved in both the GO terms encoded MHC class I molecules (β-2 microglobulin (B2 m), histocompatibility 2, K1, K region (H2-k1), histocompatibility 2, Q region locus 7 (H2-q7) and histocompatibility 2, D region locus 1 (H2-l); Tables 2 and 3).
Table 2 Up-regulated genes involved in the positive regulation of T-cell-mediated cytotoxicity
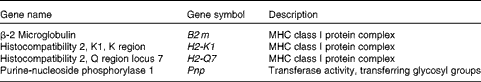
MHC, major histocompatibility complex.
Table 3 Up-regulated genes involved in the antigen processing and presentation of peptide antigen via major histocompatibility complex class I

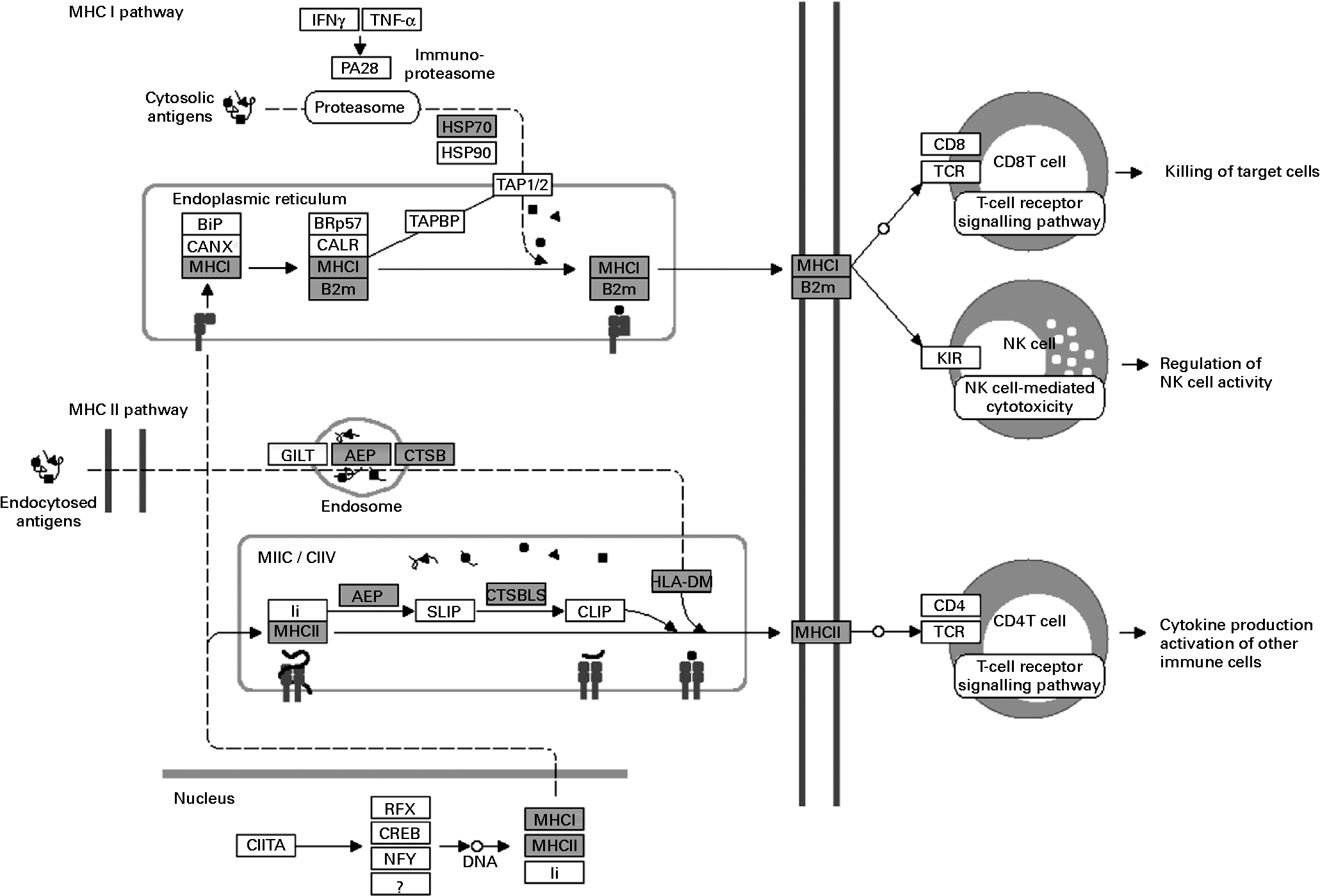
The over-represented KEGG pathways associated with cell-mediated cytotoxicity were those of NK cell-mediated cytotoxicity and those of antigen processing and presentation (Table 1). Fig. 4 shows the significantly up-regulated genes involved in the latter. Heat shock protein 1B (Hspa1b), which has chaperone activity within MHC class I, was up-regulated other than genes directly encoding MHC class I molecules. The up-regulated genes were found in the MHC class I pathway as well as in the MHC class II pathway in antigen-presenting cells. However, there were no significantly up-regulated genes encoding CD4, CD8 and T-cell receptor (TCR) composing the TCR complex.

Fig. 4 Up-regulated genes involved in antigen processing and presentation (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database; adaptation of the KEGG antigen processing and presentation pathway). Shaded boxes indicate genes significantly (false discovery rate < 0·05) up-regulated by Lactobacillus brevis KB290 feeding as revealed by DNA microarray analysis. IFN, interferon; HSP, heat shock protein; MHC, major histocompatibility complex; B2M, β-2 microglobulin; TCR, T-cell receptor; NK, natural killer. PA28, proteasome (prosome, macropain) 28; BiP, heat shock protein 5; CANX, calnexin; BRp57, protein disulfide isomerase associated 3; CALR, calreticulin; TAPBP, TAP binding protein; TAP1/2, transporter 1/2, ATP-binding cassette; sub-family B (MDR/TAP); GILT, interferon gamma inducible protein 30; AEP, legumain; CTSB, cathepsin B; MIIC, MHC class II compartments; CIIV, class II vesicles; Ii, SLIP, and CLIP, CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated); CTSB/LB, cathepsin B; HLA-DM, histocompatibility 2, class II, locus DMa; CIITA, class II transactivator; RFX, regulatory factor X-associated protein; CREB, cAMP responsive element binding protein 1; NFY, nuclear transcription factor-Y; KIR, killer cell lectin-like receptor.
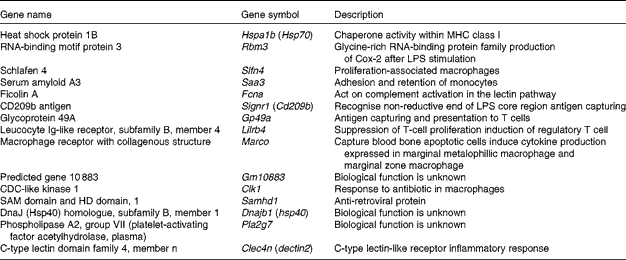
Since macrophages and DC stimulate CD8+ cytotoxic T cells via MHC class I presentation as antigen-presenting cells, the relevant up-regulated genes were selected and categorised (Table 4). Glycoprotein 49A (Gp49a), Signr1, Marco and C-type lectin domain family 4, member n (Clec4n), which act as an antigen-capturing receptor, were up-regulated.
Table 4 Up-regulated genes related to macrophages by Lactobacillus brevis KB290 (KB290) ingestion

MHC, major histocompatibility complex; LPS, lipopolysaccharide.
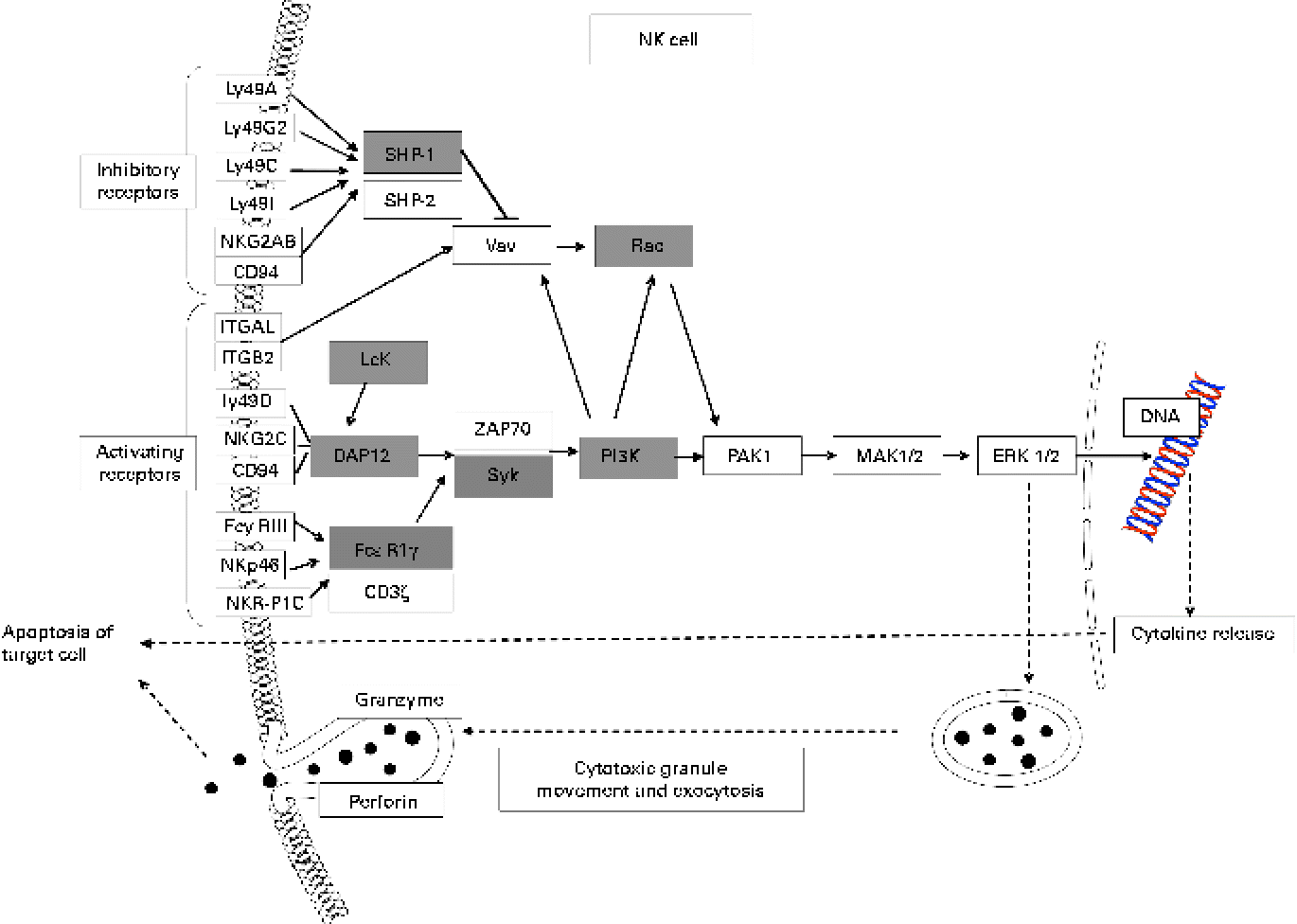
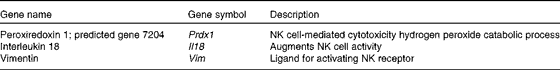
Many up-regulated genes were also implicated in NK cell-mediated cytotoxicity in the KEGG pathway and found in the NK cell activating signalling pathway, though cytotoxic molecules, such as perforin, granzyme and fibroblast-associated cell surface (FasL), were not up-regulated (Fig. 5). Moreover, the genes associated with the activation of NK cells were selected from the DEG, with the result that peroxiredoxin 1 (Prdx1), Il18 and vimentin (Vim) were found to be up-regulated (Table 5).

Fig. 5 Up-regulated genes involved in natural killer cell (NK)-mediated cytotoxicity (Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database; adaptation of the KEGG NK cell-mediated cytotoxicity pathway). Shaded boxes indicate genes significantly (false discovery rate < 0·05) up-regulated by KB290 feeding as revealed by DNA microarray analysis. Ly49a, killer cell lectin-like receptor, subfamily A, member 1; Ly49G2, killer cell lectin-like receptor, subfamily A, member 7; Ly49C, killer cell lectin-like receptor, subfamily A, member 3; Ly49I, killer cell lectin-like receptor subfamily A, member 9; NKG2AB, killer cell lectin-like receptor subfamily C, member 1; ITGAL, integrin alpha L; ITGB2, integrin beta 2; Ly49D, killer cell lectin-like receptor, subfamily A, member 4; NKG2C, killer cell lectin-like receptor subfamily C, member 2; FcyRIII, Fc receptor, IgG, low affinity IV; NKp46, natural cytotoxicity triggering receptor 1; NKR-P1C, killer cell lectin-like receptor subfamily B member 1C; SHP-1, Src homology region 2 domain-containing phosphatase 1; Vav, vav oncogene; Rac, ras-related Clck3 botulinum toxin substrate; Lck, lymphocyte-specific protein tyrosine kinase; DAP12, DNAX activation protein of 12 kDa; FcεRIγ, Fc receptor, IgE, high affinity I, gamma polypeptide; ZAP70, zeta-chain (TCR) associated protein kinase; Syk, spleen tyrosine kinase; PI3K, phosphatidylinositol 3-kinase; PAK1, p21 protein (Cdc42/Rac)-activated kinase 1; MAK1/2, mitogen-activated protein kinase kinase 1/2; ERK1/2, mitogen-activated protein kinase 1/2.
Table 5 Up-regulated genes associated with the activation of natural killer (NK) cells by Lactobacillus brevis KB290 (KB290) ingestion
Quantitative RT-PCR
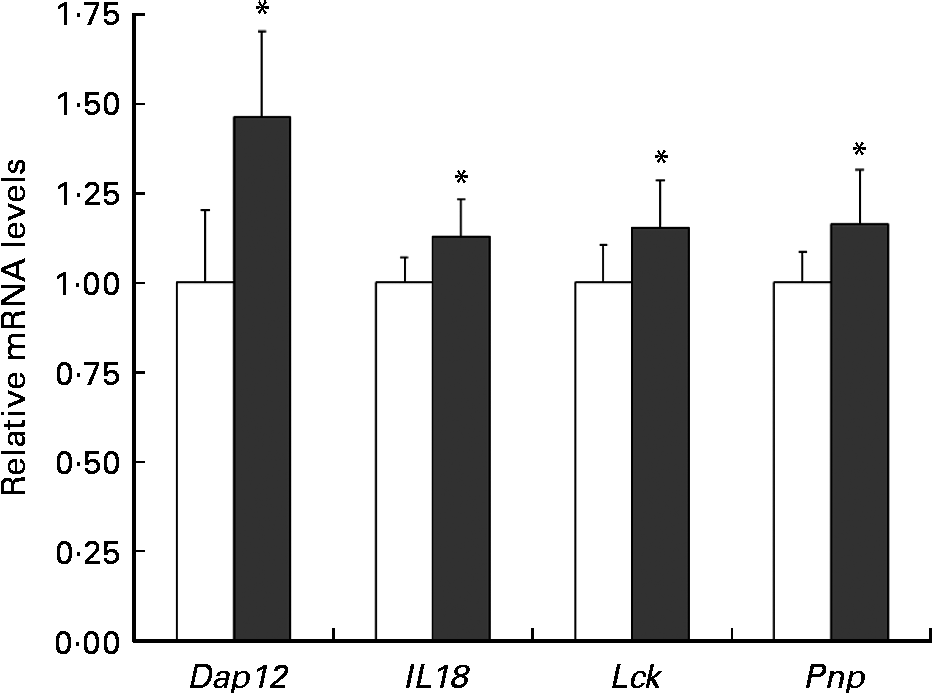
To verify the changes in gene expression detected by DNA microarray analysis, quantitative RT-PCR was applied to four selected genes implicated in the positive regulation of T-cell-mediated cytotoxicity (Pnp), the NK cell activating signalling pathway (Dap12, Lck) and the cytotoxic activity of NK cells and T cells (Il18). The results from the quantitative RT-PCR of Dap12, Il18, Lck and Pnp argued for these from the DNA microarray (Fig. 6).

Fig. 6 Relative Dap12 and Il18 mRNA levels in the spleen of mice ingested Lactobacillus brevis KB290 (KB290). The relative expression ratio (2− ΔΔCt) of each target gene in mice fed the KB290 (3 × 109 colony-forming units/g) diet (■) compared with those fed the control diet (□). The target gene expression was normalised to that of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh). Values are means, with standard deviations represented by vertical bars (n 6). * Mean values were significantly different compared with the control group (P< 0·05; Student's t test).
Proportion of marginal zone macrophages in the spleen
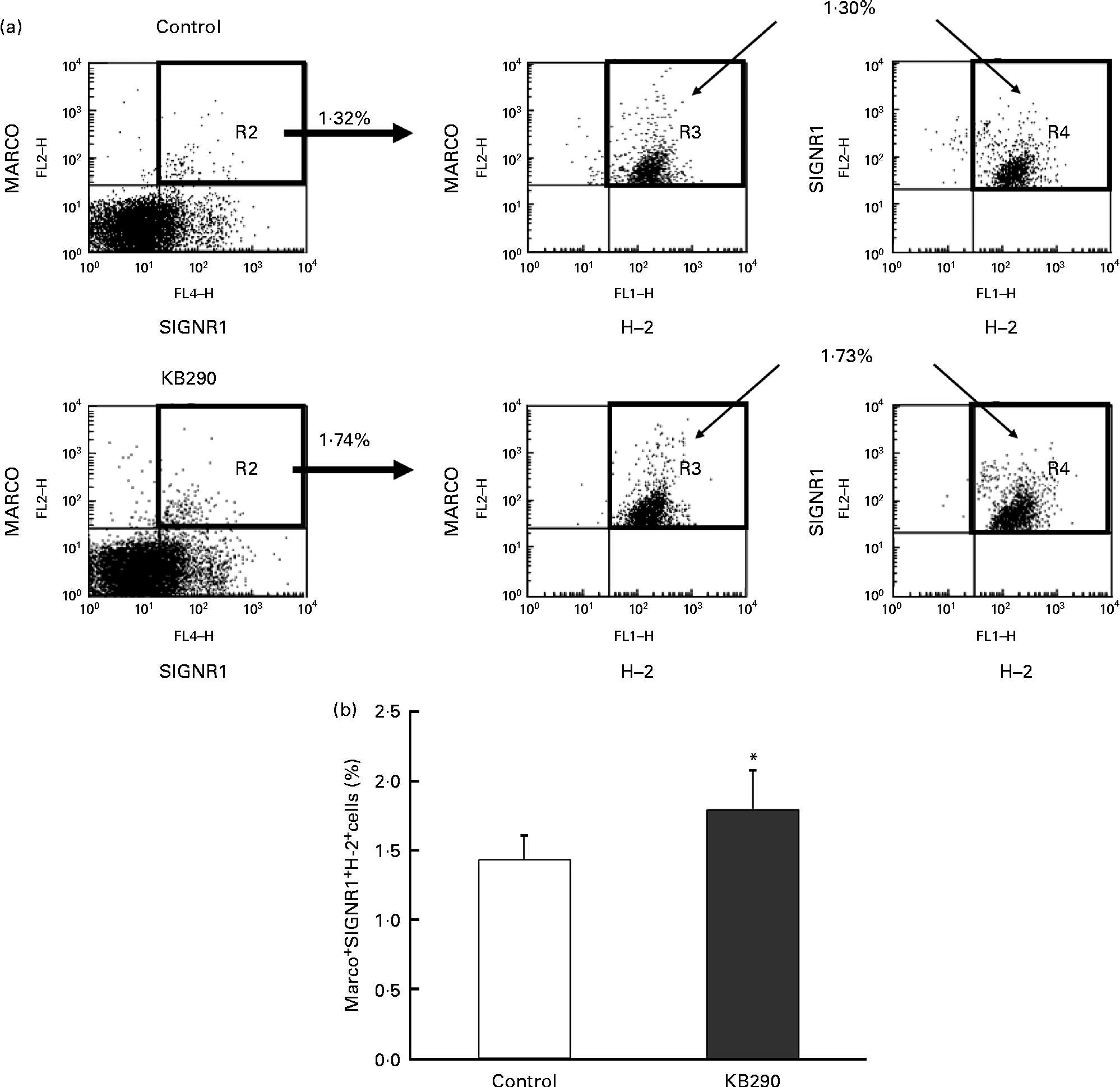
The genes encoding MHC class I molecules and the MZM-specific markers Marco and Signr1 (Table 4) were up-regulated. Therefore, the control and the treatment groups were compared for the proportion of MARCO+ SIGNR1+ H-2+ cells as MZM by flow cytometry. Typical figures for the flow cytometric analyses of splenocytes isolated from mice fed the control diet (upper panels) and the KB290 diet (lower panels) for 1 d are shown in Fig. 7(a). The proportion of MARCO+ SIGNR1+ H-2+ cells in the group fed the KB290 diet was significantly (P <0·05) increased compared with the case of the control diet (Fig. 7(b)).

Fig. 7 Effect of Lactobacillus brevis KB290 (KB290) on the proportion of marginal zone macrophages in splenocytes. Splenocytes were collected from mice fed the KB290 or control diet for 1 d and analysed for MARCO, SIGNR1 and histocompatability-2 (H-2) expression. (a) Staining with anti-MARCO and anti-H-2 is shown in the right panels for selected MARCO+ SIGNR1+ cells (as shown in the left panels). Percentage of positive cells is indicated in each quadrant. One representative sample for each group is shown. (b) The percentage of MARCO+ SIGNR1+ H-2+ cells among the total cells is shown. ■, Control group; □, KB290 group. Data from one of two experiments with similar results are shown. Values are means, with standard deviations represented by vertical bars (n 6). * Mean value was significantly different compared with the control group (P< 0·05; Student's t test). FL2-H, fluorescence channel 2 height; FL4-H, fluorescene channel 4 height; FL1-H, fluorescence channel 1 height. Fluorescein isothiocyanate was detected in FL1, R-phycoerythrin was measured in FL2-H and Alexa Fluor 647 was detected in FL4.
Discussion
It has been reported that immunomodulatory effects by probiotics are dose, administration time and strain dependent(Reference Kirjavainen, ElNezami and Salminen29, Reference Wen, Li and Bui30). Wang et al. (Reference Wang, O'Gorman and Bu31) reported that probiotics VSL#3 (11·25 × 109 bacteria/(100 mg VSL#3 × mouse), suspended in 200 μl PBS) altered the distribution of DC subsets within the intestinal mucosa after 7 d. As 1 d feeding of KB290 (1 × 1010 cfu/d) was able to induce the cell-mediated cytotoxic activity of mouse splenocytes against YAC-1 cells reproducibly (Y Fukui and E Sasaki, unpublished results), we chose the same dose and duration to investigate the relationship between the cytotoxicity of splenocytes and gene expressions related to immune functions in the present study, and the combination successfully showed cytotoxicity also. Further investigation suggested that enhancement was caused by the activation of NK cells and/or of CD8+ cytotoxic T cells stimulated via MHC class I up-regulation in antigen-presenting cells. As far as we know, the combination of a high dose and 1 d duration is a new trial to reveal the immune function of a probiotic.
Mouse spleen contains three subsets of DC(Reference Vremec and Shortman32, Reference Nakano, Yanagita and Gunn33) (myeloid (CD11c+ MHC II+ CD11b+ cells), lymphoid (CD11c+ MHC II+ CD8+ cells) and plasmacytoid (CD11c+ MHC IIlow B220+ Gr-1+ cells)) and three subsets of macrophages (F4/80+ red pulp macrophages, MARCO+ SIGLEC1+ marginal metallophilic macrophages and MARCO+ SIGNR1+ MZM)(Reference Backer, Schwandt and Greuter34). Interestingly, the MZM-specific markers Marco and Signr1 were up-regulated in the spleens of the treated mice. Furthermore, the proportion of MZM was significantly increased in the spleens of KB290-fed mice. MZM capture blood-borne bacterial antigens(Reference Aichele, Zinke and Grode35) and slightly induce an antigen-specific CD8+ cytotoxic T-cell response(Reference Backer, Schwandt and Greuter34). Also, the up-regulated genes included those which contribute to the recognition of exogenous bacteria (Clec4n (Reference Saijo and Iwakura36), Marco (Reference van der Laan, Dopp and Haworth37) and Signr1 (Reference Nagaoka, Takahara and Tanaka38)), the suppression of intracellular micro-organisms (Clk (Reference Kubori, Hyakutake and Nagai39), SAM domain and HD domain, 1 (Samhd1)(Reference Laguette, Sobhian and Casartelli40)), the cross-priming of CD8+ cytotoxic T cells (Hspa1b (Reference Calderwood, Murshid and Gong41), H2-k1 and H2-l), the activation of macrophages (schlafen 4 (Slfn4)(Reference van Zuylen, Garceau and Idris42)) and monocyte chemotaxis (human serum amyloid A 3 (Saa3)(Reference Han, Subramanian and Chan43)). Transgenic overexpression of Slfn4 in myeloid cells results in an increased number of macrophages in the liver and spleen(Reference van Zuylen, Garceau and Idris42). SAA3 acts as part of a complex matrix that increases the adhesion and retention of monocytes(Reference Han, Subramanian and Chan43). Additionally, Hspa1b (Reference Heimbach, Reznikov and Calkins44), Slfn4 (Reference van Zuylen, Garceau and Idris42) and Saa3 (Reference Meek, Eriksen and Benditt45) were up-regulated in macrophages for bacterial stimulation. The gene encoding IL18 with potent capacity to augment the cytotoxic activity of NK cells(Reference Liu, Mori and Hossain46) and T cells(Reference Nakanishi, Yoshimoto and Tsutsui47) was up-regulated. IL-18 is produced by macrophages when exposed to products such as lipopolysaccharide or oligodeoxynucleotides(Reference Nakanishi, Yoshimoto and Tsutsui47, Reference Roman, Martin-Orozco and Goodman48). These results suggest that KB290 could activate antigen-presenting cells including macrophages. Other than the up-regulated genes associated with antigen-presenting cells, Pnp involved in the positive regulation of T-cell-mediated cytotoxicity was up-regulated. PNP increases the cytotoxicity of CD8+ cytotoxic T cells(Reference Arpaia, Benveniste and Di Cristofano49). LCK is an integral component of NK cell signalling as well as the TCR signalling complex, and important for the induction of cytotoxicity. Furthermore, the genes encoding MHC class I molecules were up-regulated. Although additional molecular and physiological studies are required to define which cells may contribute to augmenting cell-mediated cytotoxic activity, circumstantial evidence suggests that CD8+ cytotoxic T cells might be possibly activated with antigen-presenting cells by KB290 feeding.
On the other hand, the responsible receptor genes for recognising antigens bound to MHC molecules, such as CD4, CD8 and TCR, were not up-regulated. The expression levels of the receptor are not always required for related cell activation. Paillard et al. (Reference Paillard, Sterkers and Vaquero50) reported that Tcr, Cd4 and Cd8 mRNA levels were not up-regulated even when T cells were activated. Thus, the activation of these stimulants might be enough to increase cell-mediated cytotoxicity, even though genes encoding the TCR complex were not up-regulated. Although further studies are required to define which cells are responsible for augmentation of cell-mediated cytotoxic activity, circumstantial evidence suggests that CD8+ cytotoxic T cells might be possibly activated with antigen-presenting cells by KB290 feeding.
KEGG analysis revealed that the up-regulated genes are enriched in NK cell-mediated cytotoxicity. NK cell function is regulated by a multitude of receptors, including the activating NK cell p46-related protein (NKp46) receptor, which also helps regulate NK cell interactions with other immune cells(Reference Moretta and Moretta51). Vimentin, which was up-regulated in the spleen of treated mice, is a direct ligand of NKp46 and can induce the cytolytic activity of NK cells(Reference Garg, Barnes and Porgador52). NKp46 non-covalently associates with the immunoreceptor tyrosine-based activation motif-containing adaptor proteins CD3ζ and/or the γ chain of fragment crystallisable receptors I (FcRI) for IgE (FcεRIγ)(Reference Walzer, Blery and Chaix53). Although we failed to detect any apparent change in Nkp46 expression, Fcεr1γ and genes encoding other signal transduction molecules, such as spleen tyrosine kinase (SYK), phosphatidylinositol 3-kinase (PI3K) and RAC, were up-regulated by KB290 ingestion. Moreover, Dap12 associated with multiple cell surface-activating receptors was also found to be up-regulated by the microarray and quantitative RT-PCR analyses. DAP12 contains an immunoreceptor tyrosine-based activation motif, which recruits SYK to trigger the cytotoxic cascade and cytokine release(Reference McVicar, Taylor and Gosselin54). Genes encoding Src homology region 2 domain-containing phosphatase 1 (SHP-1) implicated in the inhibitory pathway were up-regulated at the same time as activating signal transduction genes. SHP-1 suppresses the vav oncogene-ras-related C3 botulinium toxin substrate (VAV–RAC) pathway implicated in NK cytotoxicity through an early dephosphorylation of Vav(Reference Stebbins, Watzl and Billadeau55). RAC is downstream of not only VAV but also PI3K in the signalling pathway that activates NK cytolytic function(Reference Jiang, Zhong and Gilvary56). In considering that genes encoding PI3K and RAC were up-regulated, it is more likely that the pathway can be activated. Contrarily, perforin, granzyme B and FasL, which are cytotoxic molecules, were not up-regulated. However, it has been reported that up-regulated components of mitogen-activated protein kinase (MAPK) and cytotoxicity pathways are possibly associated with effective cytotoxicity function(Reference Wu, Dwyer and Dyer57). For these results, there is a possibility that the cytotoxic activity of NK cells was enhanced in the spleen of mice fed KB290. However, the up-regulated genes in the NK cell activating signalling pathway were involved in signalling in other cell types. LCK, PI3K and SHP-1 are components of the TCR signalling pathway (KEGG pathway). PI3K, RAC, SHP-1 and SKY are included in the B-cell receptor signalling pathway (KEGG pathway). Further studies are required to define which cells are responsible for the enhancement of cell-mediated cytotoxic activity by KB290.
DC and macrophages reside in Peyer's patches, capturing antigen shuttled through M-cells. In the lamina propria, DC extend their dendrite in the intestinal lumen, and directly sample intestinal bacteria(Reference Macdonald and Monteleone58). Therefore, these cells are important in responding to commensal bacteria in the intestine. Probiotics can induce IL-12 in lymph node DC, activating NK cells to produce interferon-γ and to increase their cytolytic potential(Reference Rizzello, Bonaccorsi and Dongarra13). On the other hand, the concentration of bacterial peptidoglycan in serum correlates with the concentration in faeces(Reference Philpott and Girardin59). Peptidoglycan stimulates innate immunity, inducing macrophages to produce cytokines. The administration of lipopolysaccharide, zymosan or Listeria monocytogenes to mice results in the up-regulation of MARCO(Reference Ito, Naito and Kobayashi60). These observations suggest that treatment with bacterial components may not only cause an increase in the percentage of MARCO-positive cells capable of participating in bacterial phagocytosis, but they also induce an increase in the amount of bacterial uptake by individual macrophages. Actually, specific markers of MZM, which capture blood-borne particles, were up-regulated in the spleens of KB290-fed mice in the present study. Some genes that contribute to phagocytosis and macrophage antigen presentation were also up-regulated on 1 d. Furthermore, the proportion of MZM by flow cytometric analysis was significantly increased by KB290 ingestion. These finding shed light on the new aspect of the immunomodulatory effect of probiotics. Further studies in gut-associated lymphoid and cardiovascular tissue are required to define whether bacterial components reach the spleen via the circulatory system or activated DC migrate to the spleen through the mesenteric lymph.
In summary, high-dose KB290 (about 1 × 1010 cfu) feeding for 1 d enhanced the cell-mediated cytotoxic activity of splenocytes. The response could be due to the activation of NK cells following stimulation, with natural cytotoxicity receptor ligand and/or CD8+ cytotoxic T cells being stimulated via MHC class I presentation. To our knowledge, the present study is the first to define the effect of ingested probiotics on changes in mouse spleen gene expression using DNA microarray analysis associated with the enhancement of cytotoxic activity. However, the present investigation covered only one tissue on day 1; gene expression changes need to be studied over time and in other tissues to uncover the mechanisms underlying the enhanced cytotoxic effect seen in animals fed KB290.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114513000767
Acknowledgements
The present study received no specific grant from any funding agency in the public but supported by Kagome Company Limited. We thank Ms M. Fukuda (Kagome Company Limited) for technical assistance and Dr M. Bloom (SciWrite Biomedical Writing and Editing Services) for professional editing. We also thank Drs A. Kishi, K. Akatani (Louis Pasteur Center for Medical Research) and H. Kodama (Kagome Company Limited) for excellent advice. The authors' responsibilities are as follows: Y. F., Y. N., K. A. and N. Y. designed the research; Y. F., E. S. and N. F. carried out the animal experiments and cell biological analysis; Y. F., E. S., T. I. and Y. N. carried out the molecular biology experiments and data analysis; Y. F., E. S., N. F. and Y. N. wrote the paper. All the authors approved the final manuscript. The authors declare that there is no conflict of interest.