Introduction
‘Heterobasidiomycetes’ is an old, taxonomically redundant name for a heterogeneous assemblage of distantly related basidiomycete groups, which share the characteristics of having predominantly septate basidia and gelatinous basidiomata. Several of these groups contain lichen-inhabiting fungi and have lately attracted much attention among lichenologists. Despite this, some of these groups have remained surprisingly poorly studied and here we will introduce a study of the lichenicolous Pucciniomycotina, focusing on the representatives presently classified in Chionosphaera. We will also review the recent rapid development in our understanding of the evolution and natural relationships of heterobasidiomycete groups, including the long overdue integration of the classifications of yeasts and filamentous taxa, and we do this with a particular focus on lichenicolous representatives.
Material and Methods
Taxon sampling
We used two different datasets for our phylogenetic analyses. The first one (dataset 1) included a representative sampling in the Pucciniomycotina (i.e. representative taxa of the 10 classes currently assigned to the group, and three lichenicolous Chionosphaera samples). In this dataset, a species in the Ustilaginomycotina (Ustilago tritici) was used as outgroup. Based on preliminary analyses of the first dataset, we used a second dataset focusing only on the Agaricostilbomycetes (dataset 2). This second sampling included representatives of the five accepted families in the Agaricostilbomycetes (i.e. Agaricostilbaceae, Chionosphaeraceae, Jianyuniaceae, Kondoaceae and Ruineniaceae: Wang et al. Reference Wang, Yurkov, Göker, Lumbsch, Leavitt, Groenewald, Theelen, Liu, Boekhout and Bai2016; Li et al. Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020). Here we included a larger number of lichenicolous Chionosphaera samples, to test the monophyly of the lineage including lichenicolous species and to focus on the systematic position and affinities of the lichenicolous taxa within the Agaricostilbomycetes. Chionosphaera is represented by C. apobasidialis, C. cuniculicola and a number of lichenicolous specimens assigned to Chionosphaera, including an isotype of C. coppinsii. Phyllozyma dimmenae was used as outgroup based on Wang et al. (Reference Wang, Yurkov, Göker, Lumbsch, Leavitt, Groenewald, Theelen, Liu, Boekhout and Bai2016). Species names, voucher information, and GenBank Accession numbers are given in Table 1.
Table 1. Sequences included in this study, either newly produced (in bold) or retrieved from GenBank. Host, specimen data and DNA extraction code are given for newly sequenced samples. An asterisk indicates one case where the ITS sequence comes from a different culture than that of nuSSU and nuLSU. Hash symbols ‘#’ indicate sequences of Crittendenia coppinsii obtained either from asymptomatic lichen specimens, or from the asymptomatic areas of lichen specimens with C. coppinsii.
Morphological studies
Herbarium specimens are deposited in K, S, TRH and UPS, and in the private collections of C. Björk, P. Diederich, J. Etayo, P. van den Boom, U. Groner, J.C. Zamora, and E. Zimmermann. Macroscopic images were captured using a Canon 40D camera with a Nikon BD Plan 10 microscope objective, StackShot (Cognisys) and Helicon Focus (HeliconSoft) for increasing the depth of field. Microscopic structures were studied on material mounted in water, 5% KOH, a mixture of phloxine B and 5% KOH, Congo red and Melzer's reagent using a Leica DMLB microscope fitted with DIC optics. Images were captured with a Leica EC3 camera and Helicon Focus.
DNA extraction, amplification and sequencing
DNA from lichenicolous specimens was extracted from either recently collected or dried herbarium material. Chionosphaera fruiting bodies were carefully sectioned and separated from the lichen thallus with a scalpel and tweezers, in order to minimize the lichen material in the DNA extraction. Approximately three to ten basidiomata were selected from each specimen for DNA extraction. Total DNA was extracted using the Qiagen DNeasy Plant Mini Kit (Qiagen, Venlo, the Netherlands), according to the manufacturer's instructions. We used three molecular markers for dataset 1: the small subunit (nuSSU), the internal transcribed spacer (ITS) and the large subunit (nuLSU) of the nuclear ribosomal DNA. For dataset 2, we used ITS and nuLSU only, to avoid introducing too much missing data since we did not obtain nuSSU sequences of the lichenicolous taxa from the present study.
We designed specific primers to selectively amplify the DNA of the new genus, avoiding that of other basidiomycetes and of the lichenized host. Suitable priming sites were identified by aligning available sequences of representatives of Chionosphaera s. lat. against sequences of other lichenicolous basidiomycetes and lichenized hosts (Lecanorales, Ascomycota), selecting conserved fragments that differed markedly between them.
General fungal primers, viz. ITS1F (Gardes & Bruns Reference Gardes and Bruns1993), ITS4 (White et al. Reference White, Bruns, Lee, Taylor, Innis, Gelfand, Sninsky and White1990) LR0R (Rehner & Samuels Reference Rehner and Samuels1994), LR3 (Vilgalys & Hester Reference Vilgalys and Hester1990), and newly designed primers (Table 2) were combined to amplify the ITS and a fragment of c. 600 bp of the nuLSU in the nuclear ribosomal DNA. Asymptomatic thalli of known hosts of C. coppinsii (Melanohalea spp.) were PCR screened for the presence of Chionosphaera. PCR amplifications were performed using IllustraTM Hot Start PCR beads (GE Healthcare Life Sciences, Pittsburg, California, USA), according to the manufacturer's instructions. PCR amplifications using the primer pair LR0R/LR3 were performed following Rehner & Samuels (Reference Rehner and Samuels1994). For the primer pairs ITS1F/ChioLSU 3-3 and ITS1F/Cc-R1, we used initial denaturing at 95 °C for 3 min, four cycles of 95 °C for 40 s, 53 °C for 40 s and 72 °C for 90 s, four cycles of 95 °C for 30 s, 50 °C for 30 s and 72 °C for 90 s, and finally 32 cycles of 95 °C for 30 s, 47 °C for 30 s and 72 °C for 90 s, with a final extension at 72 °C for 420 s. For the primer pair Cc-F1/ITS4, we used initial denaturing at 95 °C for 3 min, four cycles of 95 °C for 40 s, 52 °C for 40 s and 72 °C for 90 s, four cycles of 95 °C for 30 s, 49 °C for 30 s and 72 °C for 90 s, and finally 32 cycles of 95 °C for 30 s, 46 °C for 30 s and 72 °C for 90 s, with a final extension at 72 °C for 420 s.
Table 2. Primers newly designed for this study to selectively amplify the DNA of Crittendenia.

Before sequencing, the PCR products were purified with Exo-sap-ITTM (USB Corporation, Cleveland, Ohio, USA). The purified samples were either run on an automated sequencer (ABI Prism 377) located in the Molecular Systematic Laboratory at the Swedish Museum of Natural History, or sequenced by Macrogen (Madrid, Spain).
Multiple alignment and phylogenetic analyses
For phylogenetic analyses, sequences were aligned using MAFFT version 7 (Katoh et al. Reference Katoh, Rozewicki and Yamada2019) with the Q-INS-i algorithm. The alignments were trimmed to exclude ambiguously aligned regions using GBlocks (Castresana Reference Castresana2000), following the relaxed conditions described by Talavera & Castresana (Reference Talavera and Castresana2007). Individual marker datasets were analyzed individually by maximum likelihood bootstrapping to assess for conflicts. No incongruence was found, and data were further analyzed as two- or three-marker concatenations. Phylogenetic relationships were reconstructed using maximum likelihood (ML) and Bayesian approaches. We considered five independent partitions, nuSSU, ITS1, 5.8S, ITS2 and nuLSU, for dataset 1, and four independent partitions, ITS1, 5.8S, ITS2 and nuLSU, for dataset 2. Maximum likelihood analyses were carried out in RAxMLGUI 1.5 (Silvestro & Michalak Reference Silvestro and Michalak2012), a graphical front-end for RAxML (Stamatakis Reference Stamatakis2014). The GTRGAMMA model of nucleotide substitution was applied to all partitions because of constraints of the software RAxML. We performed a thorough ML search with a total of 100 runs and assessed node support by thorough bootstrap using 1000 bootstrap pseudoreplicates. Bayesian analyses were performed by Markov chain Monte Carlo (MCMC) sampling as implemented in the software MrBayes 3.2.6 (Ronquist et al. Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012). We selected substitution models for each of the regions using the corrected Akaike information criterion (AICc) as implemented in jModelTest 2 (Darriba et al. Reference Darriba, Taboada, Doallo and Posada2012), using full likelihood optimization and six discrete gamma categories. For dataset 1, the SYM + I + Γ was selected for the nuclear SSU rDNA, the HKY + Γ was selected for the ITS1, the SYM + I + Γ for the 5.8S, the HKY + Γ for the ITS2, and the GTR + Γ for the nuclear LSU rDNA. For dataset 2, the JC model was selected for the ITS1, the K80 + I for the 5.8S, the K80 + Γ for the ITS2, and the GTR + I + Γ for the nuclear LSU rDNA. The combined analyses treated the different regions as separate partitions with topology linked across partitions but separate model parameter values and proportional rates across partitions. For each combined dataset, three parallel runs were performed, each with five chains, four of which were incrementally heated with a temperature of 0.15. The analyses were diagnosed for convergence every 100 000 generations and were set to halt automatically when the average standard deviation of splits across runs in the last half of the analysis descended below 0.01. Every 100th tree was saved. The first 50% of each run was discarded as burn-in.
Results
Biology, phylogeny and classification of lichenicolous heterobasidiomycetes
Lichenicolous heterobasidiomycetes are now known to be very common, many are widespread, and the species are usually host-specific. This includes genera in the Pucciniomycotina (Chionosphaera, Cyphobasidium and Lichenozyma: Diederich et al. Reference Diederich, Lawrey and Ertz2018; Černajová & Škaloud Reference Černajová and Škaloud2019) and in the Tremellomycetes (Biatoropsis, Heteroacanthella, Heterocephalacria, Syzygospora s. lat. and Tremella s. lat.; Diederich et al. Reference Diederich, Lawrey and Ertz2018). Other than lichenicolous fungi, Pucciniomycotina and Tremellomycetes mainly comprise representatives with a variety of nutritional habits, including fungal and animal parasitism, plant parasitism and saprotrophy. Their diversity and evolution are still very poorly known and they may show potential co-evolutionary patterns with their hosts where host-specialization may be an important driver of speciation (Antonovics et al. Reference Antonovics, Hood and Partain2002; Refrégier et al. Reference Refrégier, Le Gac, Jabbour, Widmer, Shykoff, Yockteng, Hood and Giraud2008; Millanes et al. Reference Millanes, Truong, Westberg, Diederich and Wedin2014b; Aime et al. Reference Aime, Bell and Wilson2018).
The diversity of lichen-inhabiting heterobasidiomycetes is clearly very large but it took a surprisingly long time before the common, gall-like structures occurring on lichens were recognized as basidiomycetes. The earliest reference to these structures was by Dillenius (Reference Dillenius1742), who referred to the deformations by Biatoropsis usnearum s. lat. on Usnea as ‘small fleshy nodules closely appressed to the branches’ (orbiculos raro profert, sunt vero ii exigui carnei, ramis absque limbo arcte adnati). A number of later authors, including Acharius (Reference Acharius1795; and other references cited by Diederich & Christiansen Reference Diederich and Christiansen1994), mentioned gall-like structures on Usnea using different terminologies and interpretations, but it was not until Räsänen (Reference Räsänen1934) that they were connected to the presence of another fungus. When first described as an independent organism, Biatoropsis was believed to be an ascomycete (Räsänen Reference Räsänen1934). Most lichenicolous heterobasidiomycetes were thus overlooked, if not completely neglected, until the studies by Diederich (Reference Diederich1986, Reference Diederich1996) and Diederich & Christiansen (Reference Diederich and Christiansen1994). The first published observation of a lichenicolous Tremella species was by Coppins & James (Reference Coppins and James1979) on Violella fucata, even if this fungus remained formally unpublished until Diederich (Reference Diederich1986) published it as Tremella lichenicola. The monograph by Diederich (Reference Diederich1996) was effectively the start of a more thorough study of these fungi, including not only taxonomic descriptions of 41 new species (of the 54 species treated) but also the first discussions of putative relationships and a first serious attempt to classify them. Since Diederich's monograph, the number of newly discovered lichenicolous heterobasidiomycetes has continuously increased, currently reaching 74 species, and this number will certainly keep increasing in the future (Diederich et al. Reference Diederich, Lawrey and Ertz2018, Reference Diederich, Common, Braun, Heuchert, Millanes, Suija and Ertz2019; Diederich & Ertz Reference Diederich and Ertz2020). Before the first molecular studies, classifications including lichenicolous heterobasidiomycetes were based on a small number of morphological characters only and were often difficult to interpret. Diederich (Reference Diederich1996) had already pointed out that the taxonomic assignment of some taxa was only tentative. This was particularly the case in the genera Biatoropsis, Chionosphaera, Cystobasidium and Syzygospora, where the classification was principally based on basidium morphology and was still uncertain at that time (Diederich Reference Diederich1996). The same is true for Heteroacanthella ellipsospora, which is so far the only described lichenicolous species with acanthoid basidia (Zamora et al. Reference Zamora, Pérez-Ortega and Rico2014).
Diederich (Reference Diederich1996) studied not only sexual stages of heterobasidiomycetes, but also numerous conidia-forming species. He coined the term asteroconidia for star-shaped conidia, arising from characteristic conidiogenous cells that have been observed in both Filobasidiales (Heterocephalacria) and Tremellales (Tremella s. lat.) but, intriguingly, never in non-lichenicolous species. Diederich also observed and illustrated spores that multiplied through unicellular budding (Diederich Reference Diederich1996; figs 17 and 111), in effect the first observations of yeast-stages in lichenicolous representatives. Prillinger et al. (Reference Prillinger, Kraepelin, Lopandic, Schweigkofler, Molnar, Weigang and Dreyfuss1997) shortly after described five species of tremellalean yeasts isolated from epiphytic lichens. Zamora et al. (Reference Zamora, Millanes, Wedin, Rico and Pérez-Ortega2016) also mentioned and illustrated the germination of spores by budding in species of lichenicolous Tremella. Many, perhaps most, heterobasidiomycete groups are dimorphic, switching between a unicellular haploid yeast phase and a filamentous phase, frequently including dikaryotic hyphae and a spore-producing hymenium, in their life cycle. These phases often occur on different hosts or substrata. The switch between phases at least sometimes activates pathogenicity, although frequently the yeast stages form large masses of cells that do not harm the host in any visible way (Lin Reference Lin2009; Oberwinkler Reference Oberwinkler2017). Recently, it was shown that two groups of lichenicolous heterobasidiomycetes, Cyphobasidium spp. (Pucciniomycotina) and Tremella lethariae (Tremellales, Agaricomycotina), were able to complete their whole life cycle within the lichen thallus (Spribille et al. Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016; Tuovinen et al. Reference Tuovinen, Ekman, Thor, Vanderpool, Spribille and Johannesson2019), since yeasts, mycelium and hymenia are produced within the same lichen thallus. The life cycle of lichenicolous taxa has been studied less than non-lichenicolous heterobasidiomycetes, as lichen-inhabiting fungi are usually very difficult to grow in culture.
For many years, the lack of cultures created problems in assessing the classification of lichen-inhabiting heterobasidiomycetes, since key characters for the classification, such as the anatomy of the septal pores, required material in culture to be studied (Bandoni Reference Bandoni1984; Weiss et al. Reference Weiss, Bauer, Begerow, Agerer, Piepenbring and Blanz2004). Molecular phylogenetics, however, has increased our understanding of these fascinating fungi considerably in recent decades. Tremellalean fungi that accidentally amplified instead of the expected lichen fungus were frequent sources of error and confusion in early PCR-based lichen studies, as noted, for example, by Ekman (Reference Ekman1999). Tremellales have been later widely reported in metabarcoding studies of the lichen mycobiome (Fernández-Mendoza et al. Reference Fernández-Mendoza, Fleischhacker, Kopun, Grube and Muggia2017; Banchi et al. Reference Banchi, Stankovic, Fernández-Mendoza, Gionechetti, Pallavicini and Muggia2018). Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) introduced a set of nuclear rDNA primers designed to specifically amplify tremellalean basidiomycetes and thus the lichen host could usually be excluded from amplification experiments. Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) were the first to include lichenicolous Tremella s. lat. species in a larger phylogeny of Tremellomycetes. This study showed convincingly that the lichenicolous Tremella species form several unrelated groups within Tremellales, none of which is closely related to T. mesenterica, the type species of Tremella. Biatoropsis formed a group together with some lichenicolous Tremella species, despite the morphologically very different basidia that Biatoropsis species possess which initially caused Diederich & Christiansen (Reference Diederich and Christiansen1994) to suggest that it should be classified within Platygloeales. Utilizing these primers and the dataset produced by Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011), a number of morphologically distinct new Tremella species were later described (e.g. Millanes et al. Reference Millanes, Westberg, Wedin and Diederich2012, Reference Millanes, Diederich, Westberg, Knutsson and Wedin2014a, Reference Millanes, Diederich, Westberg, Pippola and Wedin2015; Diederich et al. Reference Diederich, Millanes and Wedin2014; Ariyawansa et al. Reference Ariyawansa, Hyde, Jayasiri, Buyck, Chethana, Dai, Dai, Daranagama, Jayawardena and Lücking2015; Zamora et al. Reference Zamora, Millanes, Wedin, Rico and Pérez-Ortega2016, Reference Zamora, Diederich, Millanes and Wedin2017, Reference Zamora, Millanes, Etayo and Wedin2018).
For a long time, the classification of filamentous and yeast-like heterobasidiomycetes unfortunately developed in parallel, as different groups of researchers traditionally focused on studies of either macrofungi or yeasts (Swann & Taylor Reference Swann and Taylor1995; Chen Reference Chen1998; Fell et al. Reference Fell, Roeijmans and Boekhout1999). This also hampered the development of a modern classification of heterobasidiomycete groups and the fulfilment of the ‘One fungus, one name’ principle, since some species had separate anamorph and teleomorph names for yeast and filamentous stages, respectively. In many cases the connection between these stages is also unknown, and often the sexual, filamentous stage seems to have been lost or reduced in frequency. Despite the previous attempts of integrated phylogenetic studies (Fell et al. Reference Fell, Boekhout, Fonseca, Scorzetti and Statzell-Tallman2000; Scorzetti et al. Reference Scorzetti, Fell, Fonseca and Statzell-Tallman2002; Sampaio Reference Sampaio, Agerer, Piepenbring and Blanz2004; Inácio et al. Reference Inácio, Portugal, Spencer-Martins and Fonseca2005; Matheny et al. Reference Matheny, Wang, Binder, Curtis, Lim, Nilsson, Hughes, Hofstetter, Ammirati and Schoch2007; Boekhout et al. Reference Boekhout, Fonseca, Sampaio, Bandoni, Kwon-Chung, Kurtzman, Fell and Boekhout2011; Wuczkowski et al. Reference Wuczkowski, Passoth, Turchetti, Andersson, Olstorpe, Laitila, Theelen, van Broock, Buzzini and Prillinger2011), it was only recently that a larger group effort resulted in the first integrated phylogenetic classification of yeasts and filamentous groups in the Tremellomycetes (Liu et al. Reference Liu, Wang, Göker, Groenewald, Kachalkin, Lumbsch, Millanes, Wedin, Yurkov and Boekhout2016). This study is a very good introduction to Tremellomycetes phylogeny (including the lichenicolous lifestyle) and could serve as a baseline for future studies focusing on the evolution of mycoparasitism in the group. It further highlights the polyphyly of Tremella and other genera, and discusses the widespread occurrence of morphologically defined but polyphyletic yeast groups within the Tremellomycetes. In the most recent phylogenetic and taxonomic studies (Liu et al. Reference Liu, Wang, Theelen, Groenewald, Bai and Boekhout2015, Reference Liu, Wang, Göker, Groenewald, Kachalkin, Lumbsch, Millanes, Wedin, Yurkov and Boekhout2016; Li et al. Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020), five orders, 17 families and 55 genera were accepted. The works by Liu et al. (Reference Liu, Wang, Theelen, Groenewald, Bai and Boekhout2015, Reference Liu, Wang, Göker, Groenewald, Kachalkin, Lumbsch, Millanes, Wedin, Yurkov and Boekhout2016) and Li et al. (Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020) represent a good framework on which to incorporate information and taxa, but many taxonomic issues remain unresolved, particularly regarding lichenicolous species.
Although the systematics and classification in the Tremellomycetes is far from stable, several molecular phylogenetic studies of lichenicolous representatives have contributed substantially to the recent progress. They have also provided valuable information on other aspects of the evolution of these fungi. An example is the evolution of the basidium morphology. Although this character was traditionally considered to be of great taxonomic importance in the group, it has now repeatedly been shown to be misleading when trying to circumscribe higher taxa. However, the morphological characteristics of the basidium are still useful to circumscribe taxa in the Tremellales at lower taxonomic levels. Millanes et al. (Reference Millanes, Diederich, Ekman and Wedin2011) studied the evolution of the ‘basidium habit’ (i.e. single basidia or basidia forming chains) and of the septation patterns in individual basidia. They found that the single basidium and the longitudinally septate basidium were the more ancestral character states, whereas catenulate basidia and other septation patterns in individual basidia were derived character states. It was also interesting to note that different basidium-related characters had evolved following different evolutionary models, for example, either a punctuated model (basidium habit) or a model of gradual evolution (basidium septation). This, together with frequent and independent transformations of the basidium in the phylogeny of the group, again indicated different evolutionary mechanisms involved in the variety of basidia observed, and pointed to a limited usefulness of basidium-related traits as taxonomic characters to characterize higher taxa in the Tremellomycetes (Millanes et al. Reference Millanes, Diederich, Ekman and Wedin2011). More recently, thorough phylogenetic studies of basidiomycetes have enabled dating the origin of both the Tremellomycetes (estimated mean value: 303 MA) and Tremellales (estimated mean value: 156 MA) (Floudas et al. Reference Floudas, Binder, Riley, Barry, Blanchette, Henrissat, Martínez, Otillar, Spatafora and Yadav2012; He et al. Reference He, Zhao, Hyde, Begerow, Kemler, Yurkov, McKenzie, Raspé, Kakishima and Sánchez-Ramírez2019).
Our phylogenies allowed us to investigate the possible influence of a joint evolutionary story between hosts and parasites in the diversification of tremellalean species, particularly in lichenicolous species (Millanes et al. Reference Millanes, Truong, Westberg, Diederich and Wedin2014b). We chose the Biatoropsis-Usnea system to explore early stages of speciation and to investigate mechanisms generating diversity and found that co-speciation was not a main evolutionary event in this system. Instead, divergence resulting from host specialization seemed to occur frequently through host-switch speciation (Millanes et al. Reference Millanes, Truong, Westberg, Diederich and Wedin2014b, Reference Millanes, Diederich, Westberg and Wedin2016a). Similarly, our earlier population haplotype and coalescent-based studies on lichenicolous Tremellales in Macaronesia revealed that host species and not geography influenced the genetic structure of Tremella lobariacearum (Werth et al. Reference Werth, Millanes, Wedin and Scheidegger2013).
In addition to the Tremellomycetes, the Pucciniomycotina is the other larger group of fungi including lichenicolous heterobasidiomycetes. It mainly comprises plant pathogens in the Pucciniales, and the rest of the group is remarkably ecologically and biologically diverse. Lichen-inhabiting species are represented only in the genera Chionosphaera, Cyphobasidium and Lichenozyma. Cyphobasidium was recently described as a result of our own phylogenetic studies that showed that the lichenicolous members of Cystobasidium (C. hypogymniicola and C. usneicola) formed a monophyletic group distinct from Cystobasidium s. str. and outside the Cystobasidiales (Millanes et al. Reference Millanes, Diederich and Wedin2016b). Cyphobasidium is characterized by having distinctive basidia that arise from a thick-walled structure, the probasidium, and by its lichenicolous occurrence on species of Hypogymnia and Usnea. It induces conspicuous gall-like structures, containing basidia, on the host lichen thalli.
Almost simultaneously with Millanes et al. (Reference Millanes, Diederich and Wedin2016b), Spribille and co-workers (Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016) discovered numerous yeast lineages from this group and recognized the new order Cyphobasidiales to accommodate them, although the systematics of this order is still unsettled (Kachalkin et al. Reference Kachalkin, Turchetti, Inácio, Carvalho, Mašínová, Pontes, Röhl, Glushakova, Akulov and Baldrian2019). Spribille et al. (Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016) found that the relative abundance of Cyphobasidium yeasts in the cortex of Bryoria tortuosa was correlated with the production of vulpinic acid in this lichen. The variable production of vulpinic acid had led in the past to the treatment of B. tortuosa and B. fremontii as distinct species, until molecular studies suggested they were conspecific (Velmala et al. Reference Velmala, Myllys, Halonen, Goward and Ahti2009). Spribille et al. (Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016) and Spribille (Reference Spribille2018) also suggested that cystobasidiomycete yeasts constituted a third component of the lichen symbiosis, particularly in the lichen family Parmeliaceae, and that they could play a role in the formation of the lichen cortex. This hypothesis, however, still remains to be tested. In a large review, Oberwinkler (Reference Oberwinkler2017) stated that ‘… these mycoparasites are dimorphic … having a haploid yeast phase as initial stage … it is a common phenomenon of yeasts that they propagate mitotically to produce yeast colonies …’ and concluded that the basidiomycete yeasts in lichen thalli are not a third component of the lichen symbiosis but the typical yeast-formed vegetative propagules so frequently produced by other mycoparasites. In a recent study, Lendemer et al. (Reference Lendemer, Keepers, Tripp, Pogoda, McCain and Kane2019) used metagenomic data to investigate the presence of Cyphobasidium across a much wider range of lichens, and observed that neither Cyphobasidium nor other cystobasidiomycete yeasts were commonly found outside lichens of the Parmeliaceae. Smith et al. (Reference Smith, Dal Grande, Muggia, Keuler, Divakar, Grewe, Schmitt, Lumbsch and Leavitt2020) also found little evidence of a widespread presence of Cyphobasidium yeasts in macrolichens, although Černajová & Škaloud (Reference Černajová and Škaloud2019) had characterized a high and widespread diversity of cystobasidiomycete yeasts associated with Cladonia. Interestingly, Černajová & Škaloud (Reference Černajová and Škaloud2019) also isolated these yeasts from ecorticate species and, in addition, were able to produce cultures of these yeasts from the medulla of some Cladonia specimens. This indicates that the yeasts are not exclusively limited to growing in the cortex. However, some species could still be part of a superficial biofilm, as suggested by Spribille (Reference Spribille2018) and Spribille et al. (Reference Spribille, Tagirdzhanova, Goyette, Tuovinen, Case and Zandberg2020). Regarding host-specialization, Mark et al. (Reference Mark, Laanisto, Guillermo Bueno, Niinemets, Keller and Scheidegger2020) recently investigated the specificity of Cyphobasidium yeasts, found in six common lichen species of Lecanoraceae, Parmeliaceae and Physciaceae, towards the lichen mycobionts, and they could not confirm a strong specialization of the yeasts, compared to that observed in the photobiont. But irrespective of their potential role or abundance, the discovery of a vast diversity of lichenicolous cystobasidiomycete yeasts associated with lichens in the Parmeliaceae (Spribille et al. Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016) raised new challenges for the taxonomic characterization of this diversity, and it is likely that Cyphobasidium and its possible role in lichens will continue to be discussed for some time.
In contrast with the recent activities focusing on Cyphobasidium, other lichenicolous taxa in the Pucciniomycotina remain comparatively poorly studied. One of the most intriguing and still poorly understood genera in the Pucciniomycotina is Chionosphaera, and we will focus the second part of this manuscript on some recent and novel results on its phylogeny and classification.
Chionosphaera was described by Cox (Reference Cox1976) based on a single species (C. apobasidialis) which is dimorphic and heterothallic, and forms characteristic white capitate synnemata-like fruiting bodies. The reniform spores are produced on holobasidia and the spores germinate by budding (i.e. have a distinct yeast phase). In nature, C. apobasidialis presumably grows associated with Cladosporium herbarum s. lat. (Dothideomycetes, Ascomycota). Cox (Reference Cox1976) included Chionosphaera in the Filobasidiales (Agaricomycotina) because of the basidium morphology that closely resembles that of Filobasidium floriforme. Oberwinkler & Bandoni (Reference Oberwinkler and Bandoni1982) later erected the new order Atractiellales (currently in Pucciniomycotina) that included, among others, the new family Chionosphaeraceae to accommodate both Chionosphaera and Stilbum.
Since the description of Chionosphaera apobasidialis, four additional species of Chionosphaera have been described: C. lichenicola Alstrup et al. (Alstrup Reference Alstrup1993), C. coppinsii P. Roberts (Roberts Reference Roberts1997), C. cuniculicola Kirschner et al. (Kirschner et al. Reference Kirschner, Begerow and Oberwinkler2001) and C. phylaciicola (Seifert & Bandoni) R. Kirschner & Oberw. (Seifert et al. Reference Seifert, Oberwinkler and Bandoni1992; Kirschner et al. Reference Kirschner, Begerow and Oberwinkler2001). Stilbum erythrinae Hansf. was further tentatively combined into Chionosphaera because its spores are thin-walled and the expected two-celled basidia with denticulate sterigmata were not observed (Kirschner & Chen Reference Kirschner and Chen2008). The genus currently includes six species, of which only C. coppinsii and C. lichenicola have a lichenicolous habit. Both C. coppinsii and C. lichenicola have clamp connections, just as C. erythrinae and C. phylaciicola, whereas C. apobasidialis and C. cuniculicola lack clamps. This, together with the differences in ecology, suggests that the genus is heterogeneous.
Based on molecular data, Bauer et al. (Reference Bauer, Begerow, Sampaio, Weiss and Oberwinkler2006) and Wang et al. (Reference Wang, Groenewald, Takashima, Theelen, Han, Liu, Boekhout and Bai2015, Reference Wang, Yurkov, Göker, Lumbsch, Leavitt, Groenewald, Theelen, Liu, Boekhout and Bai2016) placed Chionosphaeraceae in the Agaricostilbales (Agaricostilbomycetes, Pucciniomycotina). To date, only Chionosphaera apobasidialis and C. cuniculicola have been included in molecular studies (Sampaio et al. Reference Sampaio, Fell, Gadanho and Bauer1999; Kirschner et al. Reference Kirschner, Begerow and Oberwinkler2001; Bauer et al. Reference Bauer, Begerow, Sampaio, Weiss and Oberwinkler2006; Wang et al. Reference Wang, Groenewald, Takashima, Theelen, Han, Liu, Boekhout and Bai2015, Reference Wang, Yurkov, Göker, Lumbsch, Leavitt, Groenewald, Theelen, Liu, Boekhout and Bai2016) and the phylogenetic position of other species currently assigned to Chionosphaera, particularly of those with a lichenicolous habit, is uncertain. Although the life cycle of some Chionosphaera species (viz. C. apobasidialis and C. cuniculicola) has been studied in detail using cultured material, the yeast phase of the lichenicolous species (although mentioned and illustrated already by Roberts (Reference Roberts1997)) is very poorly understood. The hypothesis that these species can also complete their life cycle within the lichen thallus, as shown in other lichenicolous heterobasidiomycetes, remains untested.
Here we present recent results on the classification, diversity and biology of lichenicolous taxa currently included in Chionosphaera. Using molecular data, we investigate 1) the phylogenetic position of the lichenicolous Chionosphaera, 2) the diversity of lichenicolous species comprised in the genus, and 3) the hypothetical occurrence of Chionosphaera in asymptomatic thalli of lichenized hosts.
Phylogeny and systematics of lichenicolous Chionosphaera
Molecular results
We generated 28 new sequences (18 ITS and 10 nuLSU rDNA) that were aligned together with sequences already available in GenBank (Table 1). The combined matrix corresponding to dataset 1 contained 2501 aligned characters (nuLSU, 1–1645; ITS1, 1646–1692; 5.8S, 1693–1844; ITS2, 1845–1954; nuLSU, 1955–2501). The combined matrix corresponding to dataset 2 contained 995 aligned characters (ITS1, 1–90; 5.8S, 91–241; ITS2, 242–367; nuLSU, 368–995).
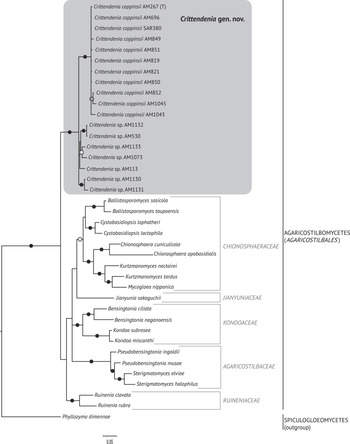
The best trees obtained from the ML analyses had lnLikelihood values of −5226.293179 for dataset 1 and −8467.292203 for dataset 2. The Bayesian analyses halted after 2 000 000 generations in analyses of dataset 1, and after 400 000 generations in analyses of dataset 2, when the average standard deviation of split frequencies across runs was < 0.01, indicating that the three runs had converged (< 0.01). In all analyses, Potential Scale Reduction Factor (PSRF) values for all model parameters as well as all branch lengths were close to 1. A majority-rule consensus tree was constructed from the 30 000 trees (dataset 1) or 6000 trees (dataset 2) of the stationary tree sample. There was no incongruence between the ML and Bayesian trees in any of the two analyzed datasets. Therefore, only the 50% majority-rule consensus tree from the Bayesian analyses corresponding to datasets 1 and 2 are shown in Figs 1 and 2, respectively, with information on ML bootstrap values added. Our analyses revealed a distinct group including all lichenicolous specimens previously assigned to Chionosphaera, placed outside Chionosphaera s. str. (Figs 1 & 2). The new genus Crittendenia is consequently described to accommodate these lichen-inhabiting taxa. Crittendenia sequences were also obtained from totally asymptomatic thalli of Melanohalea exasperatula, possibly representing an asexual yeast phase of this fungus (Table 1).
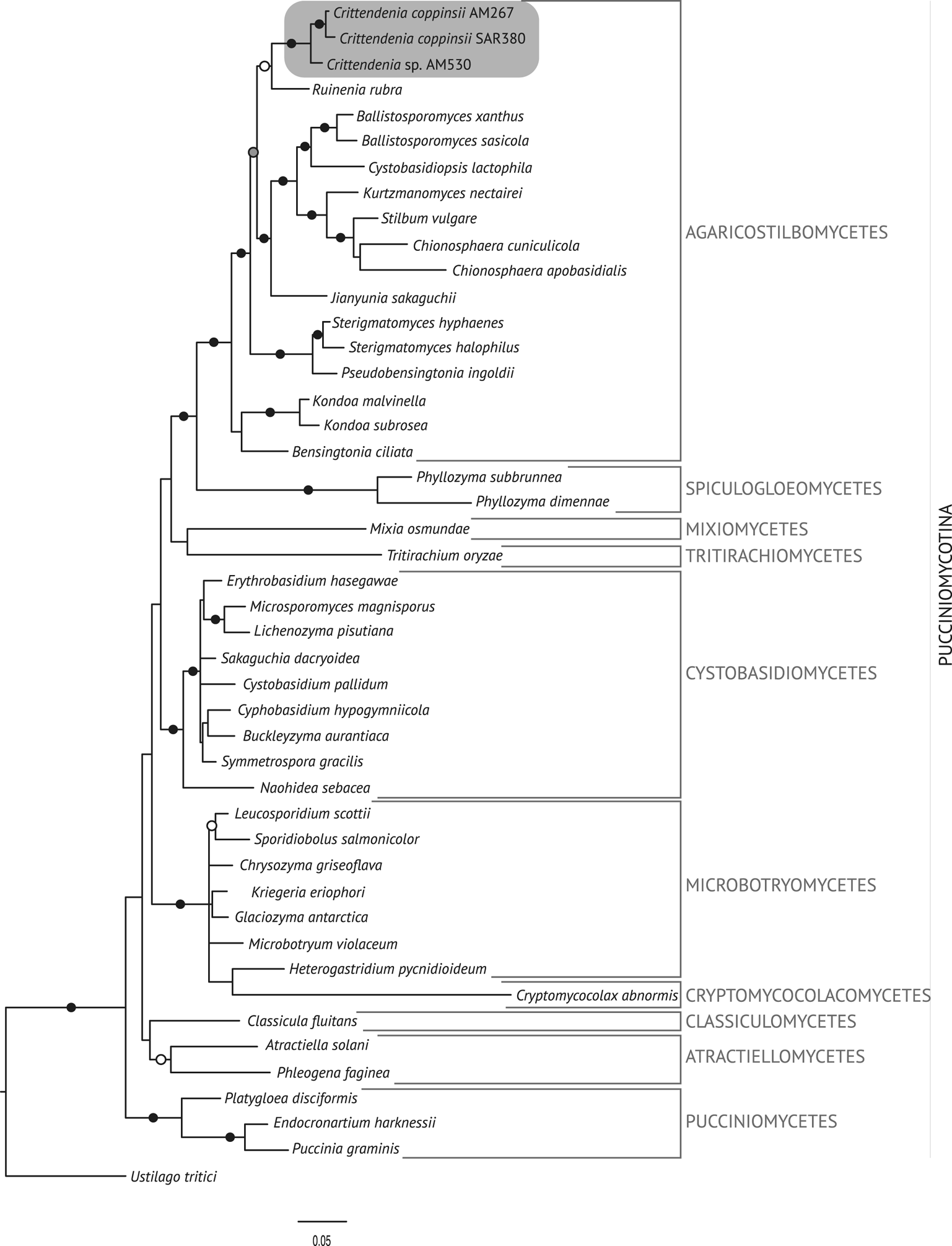
Fig. 1. Fifty percent majority-rule Bayesian consensus tree from the combined analysis including nuSSU, ITS and nuLSU, and representing the Pucciniomycotina. Black dots indicate branches supported by both Bayesian and ML analyses. White dots indicate branches supported only by Bayesian analysis. Branch lengths are scaled to the expected number of substitutions per site. Crittendenia representatives are enclosed in a grey box. Classes currently included in the Pucciniomycotina are indicated in the right margin.
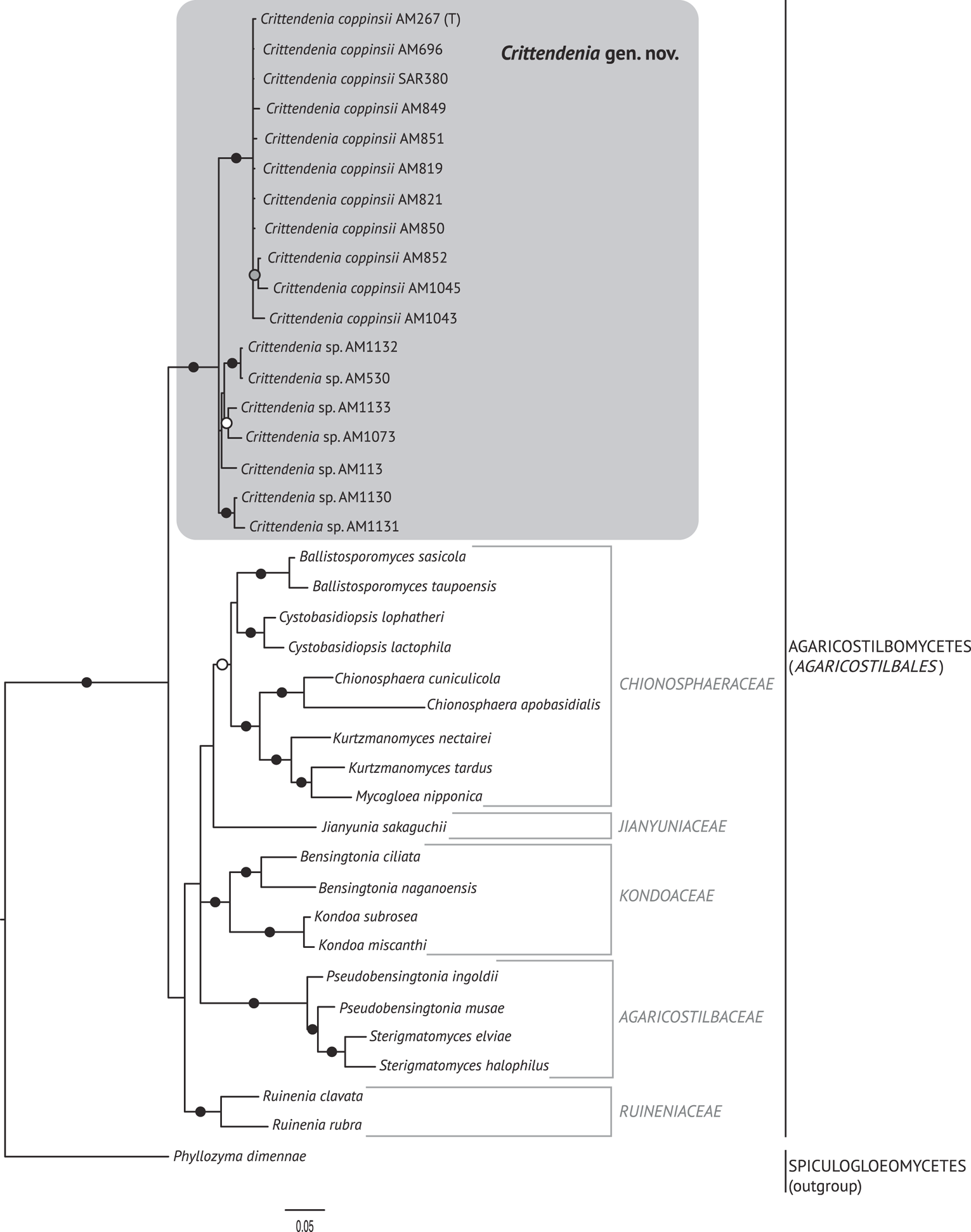
Fig. 2. Fifty percent majority rule Bayesian consensus tree from the combined analysis including ITS and nuLSU, and representing the Agaricostilbomycetes. Black dots indicate branches supported by both Bayesian and ML analyses. White and grey dots indicate branches supported only by Bayesian or ML analyses, respectively. Branch lengths are scaled to the expected number of substitutions per site. Crittendenia representatives are enclosed in a grey box and the type is indicated with ‘(T)’. Suprageneric taxa are indicated in the right margin.
Taxonomy
Crittendenia Diederich, Millanes, M. Westb., Etayo, J.C. Zamora & Wedin gen. nov.
MycoBank No.: MB 835604
Differs from Chionosphaera by the presence of basidial clamps.
Type species: Crittendenia coppinsii (P. Roberts) Diederich, M. Westb., Millanes & Wedin.
(Fig. 3)
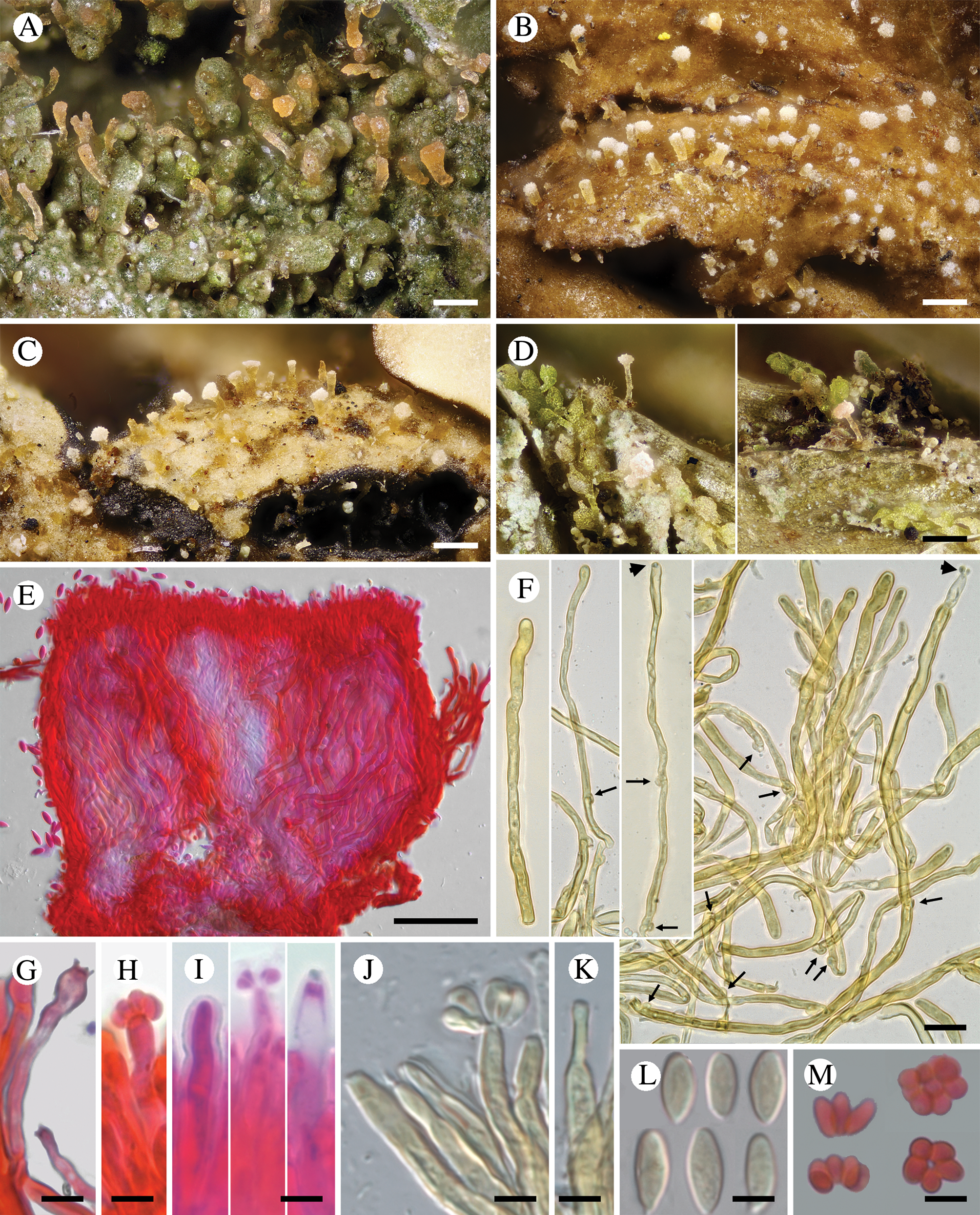
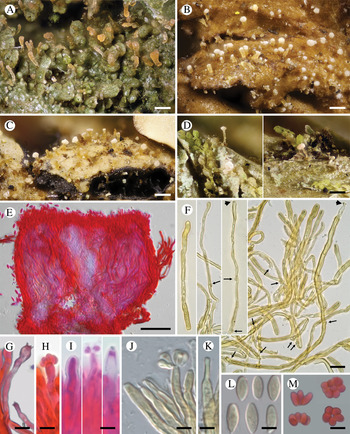
Fig. 3. Diversity of Crittendenia species. A, immature basidiomata on Melanohalea exasperatula. B, mature basidiomata on Melanelixia glabratula. C, basidiomata on Hypotrachyna laevigata. D, slender basidiomata on Fellhanera bouteillei. E, entire basidioma (in Congo red). F, basidia with basal clamps (arrows), some with inconspicuous sterigmata and/or young basidiospores (arrow heads) (in Melzer's reagent). G & H, basidia with sterigmata and basidiospores (in Congo red). I, basidium development: immature basidium (left), mature basidium with basidiospores (middle), old basidium (right) (in phloxine B). J, mature basidium with basidiospores (in Melzer's reagent). K, cystidium-like structure of unknown function (in Melzer's reagent). L, basidiospores (in Melzer). M, clusters of basidiospores separated from basidia. A, B, E, F, I–L, Crittendenia coppinsii (A & F, van den Boom 54983; B, E, I–L, Groner 714); C, Crittendenia sp. (Diederich 4913); D, Crittendenia sp. (van den Boom 56901); G, H & M, Crittendenia sp. on Bacidia (Kalb 26946). Scales: A–D = 200 μm; E = 50 μm; F = 10 μm; G–M = 5 μm.
Basidiomata developing on lichens, stipitate-capitate, synnemata-like, fleshy waxy, pale, slightly translucent; capitulum slightly to strongly differentiated and enlarged. Stipe composed of parallel, rarely branched hyphae with few septa; hyphidia and haustorial branches unknown. Basidia apical, tubular, aseptate, thin-walled, with basal clamps, when immature apically rounded, when mature with 4–7 apical, short, inconspicuous sterigmata, collapsing after spore detachment. Basidiospores hyaline, aseptate, ellipsoid to fusiform, with a small, often indistinct, basal apiculus, not forcibly discharged, often liberating together as a cluster of 4–7 spores. Basidiospores probably capable of germination by budding.
Asexual conidial stage unknown.
Etymology
We are very happy to dedicate the new genus to our friend and colleague Peter Crittenden to endorse the importance of his lichenological career. This is not only to recognize his impressive and highly valuable work as a lichen symbiosis researcher, but above all to acknowledge his outstanding contributions as Senior Editor of The Lichenologist over the past 20 years.
Ecology
Lichenicolous, associated with a large variety of lichens belonging to different phylogenetic lineages.
Crittendenia coppinsii (P. Roberts) Diederich, M. Westb., Millanes & Wedin comb. nov.
MycoBank No.: MB 835606
Basionym: Chionosphaera coppinsii P. Roberts, Mycotaxon 63, 195 (Reference Roberts1997); type: Scotland, Wester Ross, Torridon, Inveralligan, wood & gorge of Abhainn Alligin, on Melanelixia glabratula, 21 vi 1994, B. J. Coppins 16400 & A. M. O'Dare (E—holotype, non vid.; K 39188—isotype!).
A detailed description of this species is provided by Roberts (Reference Roberts1997). Crittendenia coppinsii was originally described from the UK (Scotland) and the type grows on Melanelixia glabratula (Roberts Reference Roberts1997). A second specimen provisionally reported by P. Roberts (Reference Roberts1997) and growing on Lecidella elaeochroma was later assigned to C. lichenicola by Kirschner et al. (Reference Kirschner, Begerow and Oberwinkler2001). Coppins et al. (Reference Coppins, Coppins and Douglass2009) also found C. coppinsii on a new host, Melanelixia subaurifera. Here we report Crittendenia coppinsii as new to Belgium, Norway, Sweden and Switzerland, based on specimens examined by us that grow on Melanelixia glabratula, Melanohalea exasperata and M. exasperatula. A record of ‘Chionosphaera cf. apobasidialis’ from Russia on Melanohalea olivacea is further accepted as belonging to C. coppinsii because the description fully agrees with C. coppinsii s. str. and the host is a species of Melanohalea (Zhurbenko & Himelbrant Reference Zhurbenko and Himelbrant2002). Crittendenia coppinsii should be actively searched for on other species of Melanelixia and Melanohalea.
Additional specimens examined
Belgium: Liège: Eupen, between road N68 and N620, 50°35′N, 6°2.5′E, 420 m, 2016, van den Boom 54983 (hb. van den Boom).—Norway: Møre og Romsal: Halsa, S-facing slope by Halsafjorden, Kalsetlia, 100 m, 2000, Holien 8105 (TRH); Rama, the Romsdalen Valley, S of Trollveggen Camping by the River Rauma (WP54), 6 vi 2017, Westberg & Olsson s. n. (UPS).—Sweden: Uppland: Vänge, Fiby Urskog Nature Reserve, southernmost part near entrance, 59°52.9′N, 17°21.15′E, 7 iv 2016, Westberg (S, UPS F-805352); ibid., 8 iv 2016, Westberg, Ekman & von Hirschheydt (UPS F-796396); ibid., NE of the nature reserve, 59°53.4′N, 17°21.6′E, 7 iv 2017, Westberg (S, UPS F-805353).—Switzerland: Bern: Lenk, Zelg, Simmenfälle, 1030 m, 2017, Zimmermann 1946 (hb. Zimmermann). Schwyz: Muotathal, E Fruttli, Flaschenwald, 1240 m, 1989, Groner 714 (hb. Groner).
Crittendenia lichenicola (Alstrup, B. Sutton & Tønsberg) Diederich, Millanes & Wedin comb. nov.
MycoBank No.: MB 835607
Basionym: Chionosphaera lichenicola Alstrup et al., Graphis Scripta 5, 97 (Reference Alstrup1993); type: Norway, Hordaland, Fjell, Lokøy, the peninsula S of Storafjellet, alt. 10 m, on Sorbus aucuparia, on Micarea prasina, 27 viii 1989, Tønsberg 12000 (BG—holotype!; C, IMI—isotypes!).
Descriptions of this species are provided by Alstrup (Reference Alstrup1993), Diederich (Reference Diederich1996) and Kirschner et al. (Reference Kirschner, Begerow and Oberwinkler2001). Crittendenia lichenicola differs from C. coppinsii in the much narrower and more delicate basidiomata, shorter basidia, smaller basidiospores, and host selection. Crittendenia lichenicola was originally described from Norway, growing on Micarea prasina (Alstrup Reference Alstrup1993), and has never been re-collected on this host. However, a second specimen has been published from Scotland on Micarea micrococca by Coppins & Coppins (Reference Coppins and Coppins2005). The specimen growing on Lecidella elaeochroma reported by Roberts (Reference Roberts1997) as Chionosphaera coppinsii and eventually assigned to C. lichenicola by Kirschner et al. (Reference Kirschner, Begerow and Oberwinkler2001) is here tentatively excluded from C. lichenicola, awaiting a taxonomic revision of the genus.
Discussion on Crittendenia gen. nov.
Crittendenia is a distinct and mainly lichen-inhabiting lineage in the Agaricostilbales (Pucciniomycotina), different from Chionosphaera. Crittendenia includes two known species that grow on lichen hosts of the Lecanorales: C. coppinsii is apparently confined to hosts in two closely related genera in the Parmelia-clade of the Parmeliaceae (Melanelixia and Melanohalea) (Fig. 3A & B); C. lichenicola occurs on a host of the Pilocarpaceae. Many additional specimens growing on a large variety of hosts in the families Lecanoraceae, Lobariaceae, Parmeliaceae, Physciaceae, Ramalinaceae and Teloschistaceae, some of which have been included in our phylogenetic analysis (Figs 2, 3C & D), await a morphological, taxonomic revision and are not considered further here. In a small number of cases, the interaction with a lichen host is difficult to ascertain. Since species in Crittendenia are difficult to observe in the field owing to the extremely small size of the basidiomata and the pale coloration, some of them are even difficult to detect under a binocular microscope and therefore relatively seldom collected, it is possible that the true range of hosts is even larger than reported here. Within Pucciniomycotina, Crittendenia, Cyphobasidium and Lichenozyma are the only taxa with a lichenicolous habit that have been described so far, and they are not closely related (Wang et al. Reference Wang, Yurkov, Göker, Lumbsch, Leavitt, Groenewald, Theelen, Liu, Boekhout and Bai2016; Černajová & Škaloud Reference Černajová and Škaloud2019; Li et al. Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020). Lichenozyma, isolated from Cladonia, is known only from its yeast stage, and Li et al. (Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020) suggested that it is a synonym of Microsporomyces. The latter genus would then be the only known lineage in Pucciniomycotina including both lichen-inhabiting taxa and species isolated from plant substrata. It is probable that more lichenicolous lineages are still to be discovered in Pucciniomycotina.
In addition to Chionosphaera and Crittendenia, representatives with synnematous fruiting bodies are common in Pucciniomycotina. Stilboid fruiting bodies are frequent in the Agaricostilbomycetes and are also formed in Atractiella and Phleogena (Atractiellales, Pucciniomycotina). All these taxa have, however, transversely septate basidia (Oberwinkler & Bandoni Reference Oberwinkler and Bandoni1982; Aime et al. Reference Aime, Toome, McLaughlin, McLaughlin and Spatafora2014), and none is phylogenetically closely related to Crittendenia (Fig. 1). Microscopically, the basidium of Filobasidium (Filobasidiales, Tremellomycetes) could suggest affinities to Crittendenia but Filobasidium is also phylogenetically unrelated to the new genus. Our results support existing evidence indicating that the morphology of both fruiting bodies and basidia has limited value in characterizing natural higher taxonomic groups in Basidiomycota.
Other species morphologically resembling Chionosphaera and Crittendenia, and still requiring study, are the non-lichenicolous species Chionosphaera erythrinae and C. phylaciicola (Seifert et al. Reference Seifert, Oberwinkler and Bandoni1992; Kirschner et al. Reference Kirschner, Begerow and Oberwinkler2001; Kirschner & Chen Reference Kirschner and Chen2008). Our results suggest retaining the clampless species in Chionosphaera (i.e. C. apobasidialis and C. cuniculicola), whereas those producing clamp connections (i.e. C. coppinsii and C. lichenicola) are transferred to the new genus Crittendenia (Fig. 3F). Chionosphaera erythrinae and C. phylaciicola also have clamp connections, which suggests a possible connection with the new genus Crittendenia. Chionosphaera erythrinae is known only from the type specimen that consists of two fruiting structures associated with a Cladosporium-like hyphomycete on leaves of Erythrina tomentosa. Chionosphaera phylaciicola was described from South America as Fibulostilbum phylaciicola, growing on stromata of the ascomycete Phylacia poculiformis (Seifert et al. Reference Seifert, Oberwinkler and Bandoni1992). It was later transferred to Chionosphaera by Kirschner et al. (Reference Kirschner, Begerow and Oberwinkler2001) as they considered the slight morphological differences with Chionosphaera apobasidialis, and the different fungal association, insufficient to segregate it from Chionosphaera. We unfortunately could not obtain any specimens of these two taxa for our study. Based on the host selection, the two species may be more closely related to Chionosphaera or perhaps represent different lineages, but molecular investigations will be needed to elucidate their systematic position.
The family assignment of Crittendenia within the Agaricostilbomycetes is also uncertain, until additional molecular markers other than the nuclear ribosomal DNA can be sequenced and utilized in more robust phylogenies of the group. Our analyses of the Pucciniomycotina (Fig. 1) suggest an affinity with the family Ruineniaceae. However, only the ITS and the nuLSU of the ribosomal DNA have been amplified for Crittendenia specimens. Therefore, the family allocation of the new genus should wait until a larger dataset with more markers is available for this taxon.
Our results show that the species delimitation in the new genus will need further investigation. Crittendenia coppinsii is a well-delimited species according to our phylogenetic studies and microscopical observations, and apparently grows on Melanelixia and Melanohalea only, two closely related genera in the Parmeliaceae (Blanco et al. Reference Blanco, Crespo, Divakar, Esslinger, Hawksworth and Lumbsch2004; Arup & Sandler Berlin Reference Arup and Sandler2011; Divakar et al. Reference Divakar, Crespo, Kraichak, Leavitt, Singh, Schmitt and Lumbsch2017). In contrast, herbarium specimens currently assigned to C. lichenicola probably represent a heterogeneous assemblage of several species, some of which are possibly host-specific. Host selection is an important factor that characterizes monophyletic groups in other mycoparasitic basidiomycetes (Millanes et al. Reference Millanes, Diederich, Westberg, Pippola and Wedin2015, Reference Millanes, Diederich, Westberg and Wedin2016a) and this might also be the case in Crittendenia.
Some questions still remain regarding the life cycle of Crittendenia, among them whether there is an asexual yeast phase. In addition to the observed basidiomata, we have amplified and sequenced C. coppinsii from several completely asymptomatic thalli of Melanohalea exasperatula (Table 1, Fig. 2). The yeast phase of a yet unidentified lichenicolous specimen of Crittendenia on Lecidella elaeochroma was illustrated by Roberts (Reference Roberts1997); this leads us to suspect that the sequences obtained from asymptomatic thalli correspond to yeast phases of Crittendenia, although we cannot fully exclude the presence of mycelia which, however, seem difficult to observe by light microscopy. If a yeast stage is indeed present, Crittendenia would expand the increasing quantity of previously overlooked yeast-stage diversity, recently reported from the Pucciniomycotina (Spribille et al. Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller and Thor2016; Černajová & Škaloud Reference Černajová and Škaloud2019; Kachalkin et al. Reference Kachalkin, Turchetti, Inácio, Carvalho, Mašínová, Pontes, Röhl, Glushakova, Akulov and Baldrian2019; Li et al. Reference Li, Yuan, Groenewald, Bensch, Yurkov, Li, Han, Guo, Aime and Sampaio2020).
Acknowledgements
We thank the curators of the herbaria that allowed the study of material, and Curtis Björk, Javier Etayo, Urs Groner, Pieter P. G. van den Boom, Juan Carlos Zamora and Erich Zimmermann, who kindly provided specimens for this study. The Laboratory of Molecular Systematics (MSL) at the Swedish Museum of Natural History, the Jodrell Laboratory at Kew Gardens (Kew) and the Molecular Laboratory at Rey Juan Carlos University (URJC) – in particular Bodil Cronholm (MSL), Heidi Döring (Kew), Lourdes Cano and Lidia Plaza (URJC) – are warmly thanked for excellent technical support. Heidi Döring hosted A. Millanes during a SYNTHESYS stay that allowed her to study type material. This paper was financially supported by The Swedish Taxonomy Initiative (Svenska Artprojektet, administered by the Swedish Species Information Centre/ArtDatabanken, STI dha 2016-27 4.3) and the Swedish Research Council (VR 2016-03589) through grants to M. Wedin, and by the Spanish Ministry of Economy and Competitiveness (CGL2016-80371-P) and the SYNTHESYS project GB-TAF-1326 through grants to A. Millanes.
Author ORCID
Mats Wedin, 0000-0002-8295-5198.