Introduction
With the aging of populations, the burden of neurodegenerative diseases is increasing, and substantial effort is directed toward identification of genetic biomarkers with the objective of improved disease prediction and the long-term goal of discovering therapeutic targets. In particular, molecular genetics efforts have focused on identifying common single nucleotide polymorphisms (SNPs) that contribute to disease risk.Reference Nalls, Pankratz and Lill 1 – Reference Simon-Sanchez, Schulte and Bras 3 Although these types of markers usually account for only a small proportion of disease risk, two closely linked common SNPs on chromosome 19 have been identified to jointly impart a relatively large effect on the risk of a particular neurodegenerative phenotype, namely, the apolipoprotein E (APOE) E4 allele and Alzheimer’s disease (AD).Reference Bertram, McQueen, Mullin, Blacker and Tanzi 4
APOE is found in chylomicrons, very-low-density lipoproteins, intermediate-density lipoproteins, and high-density lipoproteins; it provides structural support to these particles and also governs the catabolism of triglyceride-rich lipoproteins through its role as a receptor ligand. Importantly, APOE is the principal cholesterol carrier in the brain.Reference Mahley and Rall 5 There are three common protein isoforms of APOE – E2, E3, and E4 – historically designated based on protein mobility in isoelectrophoretic focusing gels.Reference Kane and Gowland 6 At the DNA level, these three isoforms are encoded by two nonsynonymous SNPs within the APOE gene, occurring at amino acid positions 130 and 176 (also numbered as 112 and 158, respectively, if the pro-peptide sequence is excluded), and each involving cysteine or arginine as alternate residues.Reference Zannis, Breslow and Utermann 7 The E4 allele, which has arginine at both positions 130 and 176, is the most common genetic risk factor for the development of late onset AD and contributes to disease risk in a dose-dependent manner.Reference Ward, Crean and Mercaldi 8 Meta-analyses show that one and two copies of the E4 allele raise AD risk by ~3- to 4- and ~12-fold, respectively.Reference Corder, Saunders and Strittmatter 9 , Reference Saunders, Strittmatter and Schmechel 10 Due to the replicated high-risk association from several meta-analyses of AD and APOE,Reference Ward, Crean and Mercaldi 8 , Reference Liu, Yu and Wang 11 , Reference Farrer, Cupples and Haines 12 researchers have attempted to determine whether the E4 allele is also associated with other neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS),Reference Mui, Rebeck, McKenna-Yasek, Hyman and Brown 13 , Reference Li, Pericak-Vance and Haines 14 frontotemporal dementia (FTD),Reference Agosta, Vossel and Miller 15 , Reference Geschwind, Karrim, Nelson and Miller 16 Parkinson’s disease (PD),Reference Ezquerra, Campdelacreu and Gaig 17 – Reference Huang, Chen and Poole 19 and vascular cognitive impairment (VCI),Reference Baum, Lam and Kwok 20 – Reference Chuang, Hayden and Norton 22 so far with mixed and inconsistent results.
Another gene less consistently associated with AD risk is the microtubule-associated protein tau gene (MAPT) that encodes the protein tau.Reference Myers, Kaleem and Marlowe 23 It remains to be established whether MAPT is associated with other neurodegenerative diseases. Within the MAPT gene, an ancestral inversion of ~900 kb has resulted in two distinct haplotypes, H1 and H2, and creates a large region of linkage disequilibrium. Apart from the few associations found between H1 MAPT and AD, there is debate as to whether the haplotype is associated with PDReference Seto-Salvia, Clarimon and Pagonabarraga 24 and with the FTD subtype progressive supranuclear palsy (PSP).Reference Ferrari, Wang and Vandrovcova 25 , Reference Baker, Litvan and Houlden 26
The Ontario Neurodegenerative Disease Research Initiative (ONDRI) is a multi-platform, provincial-wide, observational cohort study aiming to characterize multiple attributes of five neurodegenerative diseases, namely, (1) AD and mild cognitive impairment (AD/MCI), (2) ALS, (3) FTD, (4) PD, and (5) VCI.Reference Farhan, Bartha and Black 27 In addition to genomic analysis, ONDRI incorporates a comprehensive phenotypic assessment on each participant. The large data set, combined with the consistent enrollment criteria, allows for the unique opportunity to assess the association of APOE genotype and MAPT haplotype across the respective neurodegeneration phenotypes. Here we aim to replicate the known associations of the APOE E4 allele, APOE E4/4 genotype, and MAPT H1 haplotype with AD, in addition to assessing whether APOE E4 and MAPT H1 confer risk to ALS, FTD, PD, and VCI within the ONDRI cohort.
Methods
Blood samples were collected from 519 ONDRI participants after informed consent was obtained, in accordance with the Research Ethics Boards at Hamilton General Hospital (Hamilton, Ontario, Canada); McMaster (Hamilton, Ontario, Canada); Parkwood Hospital (London, Ontario, Canada); London Health Sciences Centre (London, Ontario, Canada); The Ottawa Hospital (Ottawa, Ontario, Canada); University Health Network- Elizabeth Bruyère Hospital (Ottawa, Ontario, Canada); Baycrest Centre for Geriatric Care (Toronto, Ontario, Canada); Centre for Addiction and Mental Health (Toronto, Ontario, Canada); St Michael’s Hospital (Toronto, Ontario, Canada); Sunnybrook Health Sciences Centre (Toronto, Ontario, Canada); and Toronto Western Hospital (Toronto, Ontario, Canada). Formal diagnoses and demographic data were obtained by participants’ clinicians upon enrollment in the study, in accordance with ONDRI standard operating protocols.Reference Farhan, Bartha and Black 27
Genomic DNA was isolated from blood samples collected from each participant, as described previously.Reference Farhan, Dilliott and Ghani 28 DNA samples were also obtained from 189 cognitively normal controls from the GenADA study.Reference Li, Wetten and Li 29 All samples underwent targeted next-generation sequencing using the ONDRISeq neurodegenerative disease gene panel. Full methodology of DNA isolation, sequencing with the ONDRISeq panel,Reference Farhan, Dilliott and Ghani 28 and raw sequencing data processing were previously described.Reference Dilliott, Farhan and Ghani 30
Allele calls for the APOE risk alleles rs429358(CT):p.Cys130Arg and rs7412(CT):p.Arg176Cys were extracted from the ONDRISeq data files and mapped to their respective APOE genotype for each participant using a customized Annotate VariationReference Wang, Li and Hakonarson 31 script. Allele calls and mapped genotypes were validated with TaqMan allelic discrimination assay,Reference Koch, Ehrenhaft and Griesser 32 as previously described.Reference Farhan, Dilliott and Ghani 28
TaqMan was also used to determine the tau haplotype of the ONDRI participants and control samples. DNA samples were genotyped for the intronic SNP rs1800547, which is not covered by the ONDRISeq panel. Based on a region of linkage disequilibrium, allele calls were mapped to their respective MAPT haplotype.Reference Lai, Bechy and Denk 33
Statistical analyses were performed using SAS v9.4 (SAS Institute, Cary, NC, USA). The Wilcoxon Mann–Whitney U test was utilized to determine the difference between the ages of the control cohort compared to the five disease cohorts of interest. Chi-square analyses were used to determine the difference between the control cohort and disease cohorts’ male–female ratios. Odds ratios (ORs) and confidence intervals (CIs) were obtained using logistic regression, adjusting for participants’ age and sex.
Results
Table 1 displays the demographics of the 519 ONDRI participants included in this study as well as the cognitively normal controls. Of the ONDRI participants, 83.0% self-reported their ethnicity as Caucasian. The ALS cohort had the lowest mean age (62.0±8.7 years), and the control cohort had the highest mean age (74.0±8.2 years), which was significantly different from the mean age of the five ONDRI disease cohorts (P<1.0×10−4). Additionally, the male–female ratio of the control cohort was significantly different from that of the overall ONDRI cohort (P<1.0×10−4).
Table 1: Demographics of the 519 ONDRI participants and 189 controls genotyped for APOE and haplotyped for MAPT
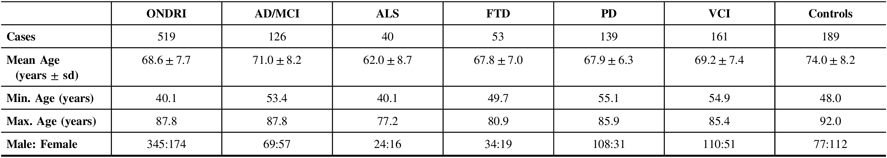
AD/MCI = Alzheimer’s disease/mild cognitive impairment; ALS = amyotrophic lateral sclerosis; APOE = apolipoprotein E gene; FTD = frontotemporal dementia; MAPT = microtubule-associated protein tau gene; Max = maximum; Min = minimum; ONDRI = Ontario Neurodegenerative Disease Research Initiative; PD = Parkinson’s disease; sd = standard deviation; VCI = vascular cognitive impairment.
Calls of the APOE alleles were obtained using the ONDRISeq panel and validated using the TaqMan allelic discrimination assay with 100% concordance. Allele and genotype frequencies were calculated for the five neurodegenerative disease cohorts and the controls (Table 2). As expected, the AD/MCI cohort displayed the highest APOE E4 allele frequency (31.7%) and E4/4 genotype frequency (14.3%), compared to 14.6% and 3.7%, respectively, in controls. The AD/MCI cohort also displayed the lowest APOE E2 allele frequency (2.4%), compared to 10.3% in controls. The lowest APOE E4 allele and E4/4 genotype frequencies were observed in the PD cohort (12.6% and 1.4%, respectively), differing marginally from the respective frequencies in controls.
Table 2: APOE allele and genotype frequencies in 519 ONDRI participants and 189 controls. All study participants were genotyped using both the ONDRISeq panel and TaqMan allelic discrimination assay
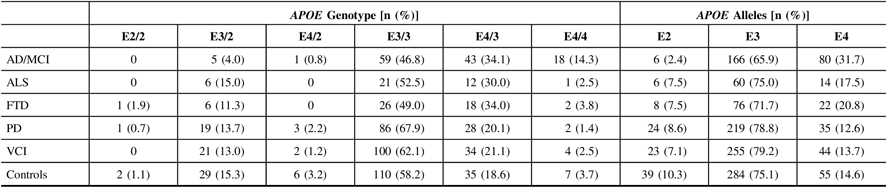
AD/MCI = Alzheimer’s disease/mild cognitive impairment; ALS = amyotrophic lateral sclerosis; APOE = apolipoprotein E gene; FTD = frontotemporal dementia; ONDRI = Ontario Neurodegenerative Disease Research Initiative; PD = Parkinson’s disease; VCI = vascular cognitive impairment.
Allele and genotype calls were compared between each ONDRI disease cohort and the control cohort, while adjusting for both age and sex of participants (Figure 1A and 1B). The E4 allele was significantly associated with increased presentation of AD/MCI compared to controls (OR=2.76, 95% CI=1.85–4.11, P<1.0×10−4). Similarly, the E4/4 genotype significantly increased the presentation of AD/MCI when compared to controls (OR=4.13, 95% CI=1.64–10.37, P=2.5×10−3). As expected, the E2 allele was associated with a significantly decreased presentation of AD/MCI when compared to controls after adjusting for age and sex (OR=0.21, 95% CI=0.08–0.50, P=5.0×10−4; Figure 2). No association with APOE was found with the other four phenotypes in the ONDRI data set.
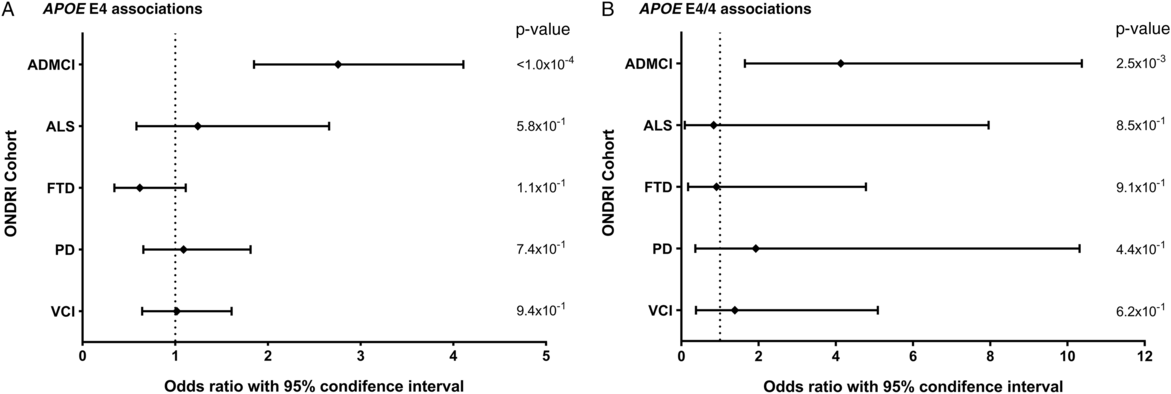
Figure 1: Forest plots of the relationship between APOE and risk of each of the diseases encompassed by ONDRI. Logistic regressions adjusting for participant age and sex analyzed the APOE E4 allele and E4/4 genotype status of the ONDRI cohorts when compared to controls. (A) Forest plot of the APOE E4 allele and associated risk of each ONDRI disease cohort. (B) Forest plot of the APOE E4/4 genotype and associated risk of each ONDRI disease cohort.
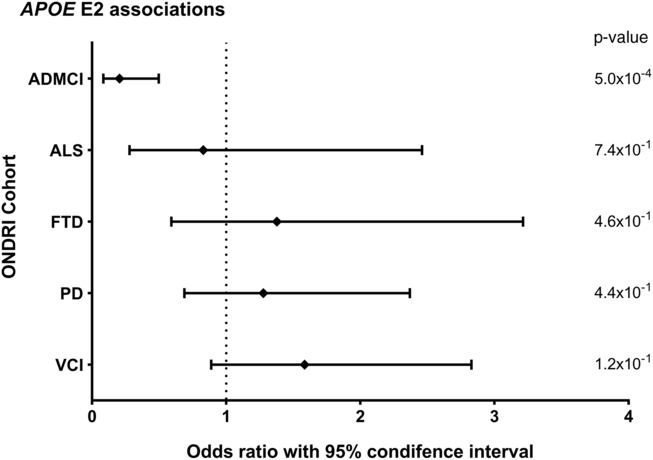
Figure 2: Forest plot of the relationship between the APOE E2 allele and risk of each of the diseases encompassed by ONDRI. Logistic regressions adjusting for participant age and sex analyzed the APOE E2 allele status of the ONDRI cohorts when compared to controls.
The AD/MCI cohort was split into participants presenting with AD (n=41) and those presenting with MCI (n=85) and APOE analyses were repeated. The AD and MCI subcohorts displayed E4 allele frequencies of 46.3% and 24.7% and E4/4 genotype frequencies of 26.8% and 8.2%, respectively. Indeed, the E4 allele was significantly associated with presentation of both AD and MCI compared to the controls (OR=5.24, 95% CI=3.07–8.92, P<1.0×10−4 and OR=1.94, 95% CI=1.22–3.07, P=4.9×10−3, respectively), and the E2 allele was significantly associated with decreased presentation of both AD and MCI compared to the controls (OR=0.10, 95% CI=0.01–0.77, P=0.0268 and OR=0.26, 95% CI=0.10–0.68, P=5.8×10−3, respectively). The E4/4 genotype was also significantly associated with increased presentation of AD (OR=10.36, 95% CI=3.55–30.19, P<1.0×10−4); however, the genotype was not significantly associated with the presentation of MCI.
Allele calls of the intronic MAPT variant, rs1800547, were mapped to their respective MAPT haplotype for each DNA sample. The ALS cohort had the highest frequencies of H1 haplotype and H1/H1 diplotype (87.5% and 75.0%, respectively), whereas the FTD cohort displayed the lowest frequencies (75.5% and 60.4%, respectively; Table 3). There were no significant associations found between MAPT and any of the disease phenotypes in ONDRI when compared to controls following adjustment for both age and sex.
Table 3: MAPT haplotype and diplotype frequencies in 519 ONDRI participants and 189 controls. All study participants were genotyped for the intronic SNP rs1800547 using TaqMan allelic discrimination assay and results were mapped to their respective haplotype
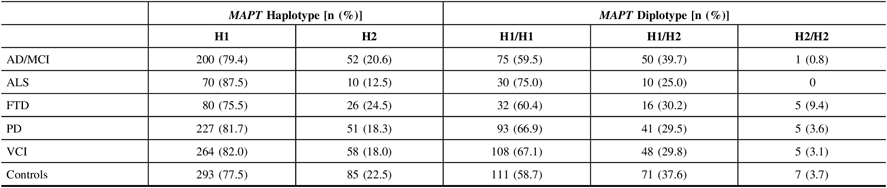
AD/MCI = Alzheimer’s disease/mild cognitive impairment; ALS = amyotrophic lateral sclerosis; FTD = frontotemporal dementia; MAPT = microtubule-associated protein tau gene; ONDRI = Ontario Neurodegenerative Disease Research Initiative; PD = Parkinson’s disease; VCI = vascular cognitive impairment.
Due to its previous associations with the PSP subtype of FTD, the FTD cohort was split into its respective subtypes, including behavioral variant FTD (bvFTD; n=22), corticobasal syndrome (CBS; n=3), progressive non-fluent aphasia (PNFA; n=8), PSP (n=15), and semantic dementia (SD; n=5), and MAPT analyses were repeated. Interestingly, the SD subcohort displayed the greatest MAPT H1 haplotype frequency and the CBS subcohort displayed the lowest, at 90.0% and 50.0%, respectively. Similarly, the SD subcohort, along with the PSP subcohort, displayed the greatest H1H1 diplotype frequency of 80.0%, while the CBS subcohort displayed the lowest of 33.3%. We also observed that the H1 haplotype was significantly associated with increased PSP prevalence (OR=7.46, 95% CI=2.39–23.29, P=5.0×10−4) following adjustment for age and sex; however, the H1H1 diplotype did not display significant associations with PSP presentation. In addition, there were no significant associations between MAPT and any of the other FTD subtypes.
Discussion
This is the first genetic characterization of the ONDRI cohort, which is important for upcoming multimodal, multi-year, prospective observational studies of the five phenotypes (e.g., APOE/MAPT-based stratification). The principal findings from the current study are as follows: (1) a dose-dependent association of the APOE E4 allele with AD and an association between E4 and MCI, (2) an inverse association of the APOE E2 allele with AD and MCI presentation, (3) a lack of associations between the APOE alleles and other neurodegenerative diseases included in the ONDRI mandate (i.e. ALS, FTD, PD, or VCI), and (4) no associations between any of the five neurodegenerative disease cohorts and the MAPT H1 haplotype, but a significant association between H1 and the PSP subtype of FTD.
Our study design offers a unique opportunity to analyze individuals with one of five neurodegenerative diseases enrolled with strict inclusion criteria and evaluated across a wide range of platforms.Reference Farhan, Bartha and Black 27 Because of this robust workflow, we can investigate the effect of the APOE alleles and genotypes and MAPT haplotypes across multiple diseases with common assessment. The control cohort had a significantly older mean age than the ONDRI disease cohorts, as well as a significantly different male: female ratio. For this reason, logistic regression was applied to obtain ORs adjusted for both the age and sex of participants.
The E4 allele frequency in previously reported AD patients is 28–37%, while in controls it is 8–14%.Reference Farrer, Cupples and Haines 12 , Reference Heffernan, Chidgey, Peng, Masters and Roberts 34 The results presented here are comparable to these literature values, with E4 allele frequencies of 31.7% and 14.5% in the AD/MCI and control cohorts, respectively. More specifically, we observed an APOE E4 allele frequency of 46.3% in individuals with AD and 24.7% in individuals with MCI. Although the E4 allele frequency was significantly increased in cohorts of MCI compared to controls, the increase is not as great as that seen in those with AD only. Interestingly, the E4 allele has been shown to be a predictive risk factor for the clinical conversion from MCI to AD,Reference Elcoroaristizabal Martin, Fernandez Martinez and Galdos Alcelay 35 – Reference Fleisher, Sowell, Taylor, Gamst, Petersen and Thal 37 which, coupled with the increased E4 allele frequency in the MCI subcohort, may indicate that a portion of the individuals enrolled in ONDRI with MCI will experience disease progression to AD. The longitudinal nature of the ONDRI study will permit follow-up of the individuals with MCI to determine whether their APOE status predicts possible progression to AD and to evaluate the phenotypic measures most severely affected by their status.
In accordance with previous literature,Reference Corder, Saunders and Strittmatter 9 we identified that APOE E4 was associated with AD in a dose-dependent manner following adjustment for age and sex. The presence of the E4 allele produced an approximately five-fold increased risk of having AD/MCI, which was marginally greater than the estimates previously found. However, we identified the E4/4 genotype to increase risk just over 10-fold, marginally lower than the commonly reported 12-fold.Reference Corder, Saunders and Strittmatter 9 , Reference Saunders, Strittmatter and Schmechel 10 The slight discrepancies are likely due to the modest number of individuals enrolled in ONDRI with AD.
Previous studies have also suggested that the E2 allele decreases the risk of AD, which we also observed with our AD/MCI cohort.Reference Farrer, Cupples and Haines 12 , Reference Corder, Saunders and Risch 38 It is hypothesized that the stability provided by the cysteine-to-arginine variant at amino acid 176 may be contributing to this protective effectReference Zhong and Weisgraber 39 and allows the isoform to more effectively clear amyloid-β,Reference Yang, Smith, Zhou, Gandy and Martins 40 protect against synaptic degeneration,Reference Dumanis, Tesoriero and Babus 41 and facilitate antioxidant activity.Reference Miyata and Smith 42 However, because of the small sample sizes within ONDRI, no individuals in the AD/MCI cohort harbored the E2/2 genotype, and precise genotypic risk associations could not be evaluated. Despite the absence of individuals with the E2/2 genotype in the AD/MCI cohort, it is expected that two copies of the E2 allele would incrementally decrease the risk of the disease in a dose-dependent manner, particularly in those with AD, but larger cohorts would be needed to validate this hypothesis.
Although an association was not observed between MAPT H1 and the total FTD cohort, we did observe an association between the haplotype and increased presentation of PSP, as has been previously identified.Reference Baker, Litvan and Houlden 26 Yet, we were not able to replicate the previously observed increased prevalence of PSP associated with the H1H1 diplotype, again possibly as a result of the modest number of individuals with PSP. Interestingly, the SD subcohort of FTD displayed the highest MAPT H1 haplotype frequency and the same H1H1 diplotype frequency as the PSP subcohort but was not significantly associated with MAPT. We expect that the very low number of individuals enrolled in ONDRI with SD may be the driving factor in the lack of association observed here and believe that further analysis into the association between MAPT H1 and SD is warranted with larger sample sizes.
Due to the strong association between both the E4 allele and E4/4 genotype and AD status, many studies have attempted to identify associations with other neurodegenerative disorders.Reference Verghese, Castellano and Holtzman 43 However, these studies reported inconsistent results regarding the risk associated with APOE E4 and onset and/or progression of the other four diseases studied in ONDRI, namely, ALS,Reference Mui, Rebeck, McKenna-Yasek, Hyman and Brown 13 , Reference Li, Pericak-Vance and Haines 14 FTD,Reference Agosta, Vossel and Miller 15 , Reference Geschwind, Karrim, Nelson and Miller 16 PD,Reference Ezquerra, Campdelacreu and Gaig 17 – Reference Huang, Chen and Poole 19 and VCI.Reference Baum, Lam and Kwok 20 – Reference Chuang, Hayden and Norton 22 Within the ONDRI cohort, no associations were identified for the other disease phenotypes. Similarly, no associations were identified between the MAPT H1 haplotype or H1/H1 diplotype and any of the five complete neurodegenerative disease cohorts. Absence of associations could have been due to small sample sizes, and thus false negative inferences, or to the true lack of a biological effect of E4 and H1 in these conditions. Associations previously reported may have been due to the diagnostic challenges associated with neurodegenerative diseases. Admixture of AD pathology in individuals with other neurodegenerative diseases may produce false positive associations with E4, and copathologies within neurodegenerative diseases are far more common than previously appreciated.Reference Robinson, Lee and Xie 44 Due to the spectrum of overlapping features that can be observed across neurodegenerative phenotypes, it will be important to identify those that are associated with the APOE E4 allele and MAPT H1 haplotype to better understand patient prognosis. Future analyses will utilize ONDRI’s robust assessment of structural and cognitive measures to identify whether common phenotypes across the various diseases are influenced by APOE and MAPT.
While ONDRI is unique in terms of the number of different clinical conditions evaluated simultaneously within the process, there is still a limitation due to modest sample sizes. Larger cohorts may produce results that align more closely with those previously reported. Additionally, an important limitation to this study is the lack of correlation with cognitive status within the disease cohorts. Assessments of cognitive impairment are ongoing, and future studies will incorporate these measures from the participants in order to assess the effects of the APOE E4 allele and MAPT haplotype on cognitive status within all five disease cohorts.
In conclusion, E4 allele carriers in the ONDRI study displayed a dose-dependent increased risk of AD/MCI, specifically in those diagnosed with AD, which is consistent with the current APOE literature. Similarly, this study was concordant with recent evidence that the APOE E2 allele decreases the risk of AD/MCI. Further, the MAPT H1 haplotype was significantly associated with the PSP subtype of FTD. The work also confirmed that risks of the other four diseases evaluated within ONDRI, namely, ALS, FTD, PD, and VCI are not associated with the E4 allele or E4/4 genotype and that none of the complete neurodegenerative disease cohorts are associated with the MAPT H1 haplotype or H1/H1 diplotype. To our knowledge, this is the first study to analyze APOE genotypes and MAPT haplotypes across these five neurodegenerative diseases using common enrollment criteria and comprehensive phenotypic analysis. Future studies will investigate the structural and cognitive symptoms of neurodegeneration influenced by the E4 allele and H1 haplotype and the contributions of other genetic factors to these phenotypes.
Acknowledgments
We would like to thank all ONDRI participants for their consent and cooperation with our study. Thank you to the ONDRI investigators (www.ONDRI.ca/people), and the ONDRI governing committees: the executive committee, steering committee, publication committee, recruiting committee, assessment platforms, and project management team. We also thank the London Regional Genomics Centre for their technical expertise. AAD is supported by the Alzheimer Society of London and Middlesex Doctoral Graduate Research Scholarship. SMKF is supported by the ALS Canada Tim E. Noël Postdoctoral Fellowship. MF receives support from the Saul A. Silverman Family Foundation as a Canada International Scientific Exchange Program and the Morris Kerzner Memorial Fund.
Funding
This work is funded by the Ontario Neurodegenerative Disease Research Initiative, through the Ontario Brain Institute, an independent nonprofit corporation, funded partially by the Ontario government. The opinions, results, and conclusions are those of the authors and no endorsement by the Ontario Brain Institute is intended or should be inferred.
Disclosures
Dr. Masellis reports grants from Canadian Institutes of Health Research, during the conduct of the study; other from Associate Editor, Current Pharmacogenomics and Personalized Medicine, personal fees from Bioscape Medical Imaging CRO, personal fees from GE Healthcare, personal fees from UCB, grants from Canadian Institutes of Health Research, grants from Early Researcher Award, Ministry of Economic Development and Innovation of Ontario, grants from Ontario Brain Institute, grants from Sunnybrook AFP Innovation Fund, grants from Alzheimer’s Drug Discovery Foundation (ADDF), grants from Brain Canada, grants from Heart and Stroke Foundation Centre for Stroke Recovery, grants from Weston Brain Institute, personal fees from Novartis, personal fees from Henry Stewart Talks, other from Novartis, grants from Roche, grants from Washington University, grants from Teva, other from Teva, outside the submitted work. Outside the submitted work, Dr. Black reports institutional grants from GE Healthcare, Eli Lilly, Biogen Idec, Novartis, Genentech, Optina, Roche, and personal fees from Pfizer, Eli Lilly, Novartis, Merck, Medscape, Roche and Biogen Idec. Dr. Freedman reports grants from Ontario Brain Institute, during the conduct of the study. In addition, Dr. Freedman has a patent “Systems and methods for non-invasive biomarker assessment of dementia” pending. Dr. Lang reports personal fees from AbbVie, personal fees from Acorda, personal fees from Bristol-Myers Squib, personal fees from Biogen, personal fees from Merck, personal fees from Sun Pharma, personal fees from Corticobasal Solutions, personal fees from Sunovion, personal fees from Paladin, personal fees from Medichem, personal fees from Medtronic, outside the submitted work. Dr. Shoesmith reports grants from Ontario Brain Institute, during the conduct of the study. Dr. Swartz reports personal fees from The Ontario Brain Institute, during the conduct of the study.
Statement of authorship
All authors have approved this manuscript.







