1. Introduction
Bacterial blight (BB) disease of rice caused by X. oryzae pv. oryzae (Xoo) limits rice yield in all major rice-growing regions of the world especially in irrigated lowland and rainfed conditions. The estimated yield loss due to BB under severe infection varies from 50 to 80% in tropical Asia (Khush & Ogawa, Reference Khush and Ogawa1989). Deployment of resistant cultivars is the most economical and effective method to control the disease due to the non-availability of effective bactericidal agents. Two types of resistance, vertical and horizontal resistance, have been recognized to be in operation in rice against Xoo. Vertical resistance, governed by single major genes, is race specific and can be broken down easily unless two or more genes are deployed in conjunction as gene-pyramids. Through classical genetic analysis about 34 major genes conferring resistance to various strains of Xoo (Chen et al., Reference Chen, Liu, Zeng, Ouyang, Yang and Zhu2011) have been identified and most of these have been mapped to various chromosomes (Jin et al., Reference Jin, Wang, Yang, Jiang, Fan and Liu2007). So far, six BB resistance genes, Xa21, Xa1, Xa26, Xa27, xa5 and xa13 have been cloned by map-based cloning strategy (Chu et al., Reference Chu, Yuan, Yao, Ge, Yuan, Xu, Li, Fu, Li, Bennetzen, Zhang and Wang2006b). Horizontal resistance on the other hand is quantitative, presumably non-race specific and controlled by quantitative trait loci (QTLs). QTL mapping provides an effective approach to study complex and polygenic forms of disease resistance. Recent progress in the QTL analysis using DNA markers especially simple sequence repeats (SSRs) allows a better understanding of the traits that are controlled by multiple genes. QTLs responsible for the BB resistance have been identified in earlier studies (Li et al., Reference Li, Luo, Mei, Paterson and Zhao1999).
Large-scale cultivation of varieties with single resistance genes like Xa3 and Xa4 resulted in development of more virulent races making them ineffective (Shi et al., Reference Shi, Wang and Guo2001). Evolution of virulent populations of Xoo against the major resistance gene, Xa21 has been reported in some parts of India and Korea (Sirisha et al., Reference Sirisha, Reddy, Mishra, Das, Bernardo, Vera Cruz, Leung and Sridhar2004). Widespread and repeated use of a few resistance genes has accelerated the selection of new pathogenic races at a rate equivalent to 1·64 times the increase in specific virulence of the isolate following host-plant selection pressure after a single crop cycle (Nayak, Reference Nayak1986b). One of the strategies to delay breakdown of resistance is to identify new resistance genes from diverse sources especially identified broad spectrum durable resistant cultivars and to introgress them into susceptible but otherwise popular varieties.
Several varieties with resistance to BB have been released under the auspices of the All India Coordinated Rice improvement project (AICRIP) in India. The variety, Ajaya (IET 8585) was released by AICRIP in 1992 and possesses high level of resistance against most of the pathotypes in India (DRR Progress Report 1996–2009). Ajaya is commonly used as a differential cultivar and as a resistant check in AICRIP trials. However, information regarding the inheritance of BB resistance in Ajaya is not clear with one study indicating the action of two independently segregating dominant genes (Saini et al., Reference Saini, Goel and Sharma1996), while another reported the action of a single recessive gene (Kameswara Rao et al., Reference Kameswara Rao, Jena and Lakshminarasu2003). Hence, an effort was made through the present study to clarify the inheritance of resistance to BB in Ajaya, tag and map the gene(s) conferring BB resistance with the help of SSR markers and identify putative candidate genes responsible for BB resistance through an in silico approach.
2. Materials and methods
(i) Materials
The plant materials analysed in the present study consisted of the BB resistant cultivar Ajaya, one of its parents – BJ1, the BB susceptible varieties TN1, BPT 5204 (Samba Mahsuri), near isogenic lines (NILs) of IR24 possessing single BB resistance genes-xa5 (IRBB5), xa13 (IRBB13), Xa21 (IRBB21), NILs possessing combinations of Xa4, xa5, xa13 and Xa21 and the BB differential BJ1. Crosses were made with Ajaya as donor and TN1, BPT 5204, IRBB5 and IRBB13 as recipient parents to develop F2 and F3 mapping populations. A population consisting of 800 F2 individuals derived from the cross Ajaya/TN1 was used to study the inheritance of resistance to BB and for mapping the resistance gene(s) in Ajaya. An alternate population consisting of 592 F2 individuals obtained from the cross Ajaya/BPT 5204 was used for validation of genetic distances of the linked markers. Two sets of populations, each consisting of 177 F2 individuals derived from Ajaya/IRBB5 and 326 F2 individuals of Ajaya/IRBB13 were used for allelism test of xa5 and xa13, respectively. Seven strains of Xoo, collected from different hotspot locations of India were used to study the level and spectrum of BB resistance of Ajaya along with NILs possessing single or combination of BB resistance genes and susceptible parents (Table 1).
Table 1. Disease reaction of Ajaya along with NILs of IR24 with seven hypervirulent isolates of Xoo

Note: DX-066 collected from Raipur (Chattisgarh); DX-002 collected from Faizabad (Uttar Pradesh); DX-018 collected from Kapurtala (Punjab); DX-084 collected from Kaul (Haryana); DX-109 collected from Kerala; DX-119 collected from Hyderabad (Andhra Pradesh); DX-139 collected from Sambalpur (Orissa).
(ii) Methods
(a) Disease evaluation
The parents, F1, F2 and F3 populations of the above crosses were phenotyped for their reaction to BB at 55 days after sowing which coincides with the active tillering phase of the crop. The bacterial culture was grown on modified Wakimoto's medium (sucrose 20·0 g; peptone 5·0 g; Ca(NO3)2, 4H2O 0·5 g; Na2HPO4 0·82 g; FeSO4, 7H2O 0·05 g; agar 20·0 g; distilled water 1000 ml; pH 6·8–7·0) for 3 days at 28°C. The bacterial cells were suspended in sterile distilled water such that approximately a concentration of 108–109 cfu/ml was maintained. The parents and the mapping populations were inoculated with the bacterial suspension by the leaf clipping method (Kauffman et al., Reference Kauffman, Reddy, Hsieh and Merca1973). Plant reaction to the BB pathogen was recorded 14 days after inoculation. Plants were scored based on the per cent diseased leaf area (DLA) following the conditions described by Chen et al. (Reference Chen, Lin, Xu and Zhang2000) and IRRI–SES system of scoring (Anonymous, 2002). The scores from 5 to 6 inoculated leaves from a single F2 plant were averaged and categorized as highly resistant (score 1, % DLA<5), moderately resistant (score 3, % DLA=5–11·9), moderately susceptible (score 5, % DLA=12–24·9), susceptible (score 7, % DLA=25–49·9) and highly susceptible (score 9, % DLA=50–100).
(b) PCR analysis
Total genomic DNA was extracted from the parents and mapping populations derived from the crosses Ajaya/TN1 and Ajaya/BPT 5204 using the procedure of Zheng et al. (Reference Zheng, Huang, Bennett and Khush1995) . PCR was performed in a thermal cycler (Perkin–Elmer 480, USA) as per the conditions described by Chen et al. (Reference Chen, Temnykh, Xu, Cho and McCouch1997) . SSR marker resolution was done on 4% agarose gels stained with ethidium bromide and photographed under ultraviolet light. In order to test the presence of xa5 and xa13 in Ajaya, the xa5-linked CAPS marker RG556 (Huang et al., Reference Huang, Angeles, Domingo, Magpantay, Singh, Zhang, Kumaravadivel, Bennett and Khush1997) and the xa13-linked PCR-based marker xa13-prom (Chu et al., Reference Chu, Yuan, Yao, Ge, Yuan, Xu, Li, Fu, Li, Bennetzen, Zhang and Wang2006b) were analysed in Ajaya along with IR24, IRBB5, IRBB13, TN1, BPT 5204 and the parental line BJ1. Restriction digestion for the CAPS marker RG556 was performed in a reaction mixture consisting of 3·2 μl sterile distilled water, 1·5 μl restriction buffer (10×), 0·3 μl restriction enzyme Dra 1 (3U/μl) and 20 μl PCR product by incubating at 37°C for 1 h. The samples were then resolved in a 2% agarose gel and visualized after staining with ethidium bromide. PCR analysis of xa13prom was done following conditions similar to SSRs and the amplicons were resolved in 2% agarose gel.
(c) Analysis of functional nucleotide polymorphism with respect to xa5 in Ajaya
A primer pair was designed based on a region on chromosome 5 of Nipponbare rice sequence (Pseudo molecule5) from 415 388 to 415 630 bp, which encompasses the 2 bp functional polymorphism TC/AG in the exon II of the transcription factor IIAγ (Iyer & McCouch, Reference Iyer and McCouch2004). The primers were used to amplify the resistant and susceptible genotypes (IRBB5, Ajaya, TN1 and BPT 5204). The primer sequences were as follows: xa5F: TCC CCC CAC CCC AAA AAG, xa5R: ACA AAC AAA ATG AGG AGT CG. PCR amplification was performed using a high-fidelity DNA polymerase–Pfu polymerase (Promega, USA) to avoid errors as per the conditions described earlier. The PCR products were resolved on a 0·8% agarose gel and eluted using Wizard ®SV Gel and PCR clean-up system (Promega, USA). The products were cloned using Qiagen® PCR cloning kit (Qiagen, Germany) as per the manufacturer's instructions. Plasmids were isolated from the transformed colonies using Qiaquick plasmid isolation kit (Qiagen, Germany) and sequenced in both directions in an ABI prism 3700 automated DNA sequencer using the T7 and T13 primers of the vector. Vector sequences were removed from the sequenced fragments and the cleaned-up fragments were then aligned using Clustal W (available at the website http://www.ebi.ac.uk/clustalw) program of the European Bioinformatics Institute (Thompson et al., Reference Thompson, Higgins and Gibson1994).
(d) Molecular mapping using SSR markers
Bulked segregant analysis (BSA)
In order to map the BB resistance genes in Ajaya and to identify linked SSR markers, a set of 480 SSR markers spread across the rice genome were used. BSA was carried out by amplification of the resistant and susceptible parents along with the RB and SB with the polymorphic SSR markers. Markers that displayed bulk-specific amplification were analysed in the entire mapping population to study their linkage distance with respect to the resistance genes.
Fine mapping
Once the tentative chromosomal location of the resistance loci was known through BSA, all the SSR markers in the vicinity, possessing a repeat length of more than 20 bp, were selected for genotyping the F2 population of Ajaya/TN1 for coarse mapping. Fine-mapping of the two putative resistance-associated loci was carried out using a separate set of 22 SSR markers specific to chromosome 5 (in the vicinity of xa5) and 45 SSR markers specific to the long arm of chromosome 8. Of the 45 markers, 39 were from the RM series (available at the website www.gramene.org) and six markers viz., RMAFM1 to RMAFM6 (see Supplementary Table S1 available at http://journals.cambridge.org/grh) were specifically designed by FASTPCR software using the flanking sequence data of the repeat motifs identified in-between the flanking markers by SSRIT tool of Gramene (available at the website http://www.gramene.org). Linkage analysis and map construction were performed using MAPMAKER/EXP, version 3 (Lander et al., Reference Lander, Green, Abrahamson, Barlow, Daly, Lincoln and Newburg1987). QTL analysis was done using Composite interval mapping function of Windows QTL Cartographer ver 2.5 (Zeng, Reference Zeng1994) using per cent DLA as trait phenotype.
(e) In silico physical mapping of BB resistance gene
The physical map of the resistance gene was constructed by bioinformatics analysis using bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) clones of cv. Nipponbare released by the International Rice Genome Sequencing Project (IRGSP, 2005). The flanking markers were assigned to the respective BAC or PAC clones through BLASTN analysis. The PAC/BAC clones were aligned using the software tool Pairwise BLAST (available at the website http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html).
(f) Candidate gene annotation
Based on the targeted region of the resistance gene, the publicly available BAC or PAC sequences of Oryza sativa cv. Nipponbare were analysed by gene prediction programmes FGENESH (available at the website http://genomic.sanger.ac.uk), Rice GAAS (available at the website http://ricegaas.dna.affrc.go.jp/) and Gene Scan (available at the website http://genes.mit.edu/GENSCAN). The candidate genes were analysed through BLAST (available at the website http://www.ncbi.nlm.nih.gov/blast/) and confirmed by the TIGR Rice Genome Annotation Version 6.0 (available at the website http://rice.plantbiology.msu.edu/cgi-bin/gbrowse/rice/).
3. Results
(i) Genetic analysis of BB resistance in Ajaya
When screened with multiple isolates of Xoo, Ajaya displayed a broader spectrum of resistance as compared with the susceptible checks (TN1 and BPT 5204) and the NILs possessing the single resistance genes, Xa4, xa5, xa13 and Xa21 (Table 1). The resistant parent Ajaya and the susceptible parents TN1 and BPT 5204 showed clear distinction in terms of disease score with Ajaya displaying a score of 1 and the susceptible parents, TN1 and BPT 5204 displaying a score of 9. The F1s were moderately susceptible with a disease score of 5. Out of the 800 F2 plants screened, 48 were highly resistant, 210 were moderately resistant, 274 were moderately susceptible, 212 were susceptible and 56 were highly susceptible (Fig. 1(A), see Supplementary Table S2 available at http://journals.cambridge.org/grh). This fitted well in a segregation ratio of 1:4:6:4:1 (χ2=4·27, P=0·37) with respect to high resistance, moderate resistance, moderate susceptibility, susceptibility and high susceptibility. Similar segregation pattern was observed in the F2 population derived from the cross Ajaya/BPT 5204 (Fig. 1(B), see Supplementary Table S3 available at http://journals.cambridge.org/grh). The F1s were moderately susceptible (score 5) and among the 592 F2 plants, 32 were resistant, 139 were moderately resistant, 235 were moderately susceptible, 146 were susceptible and 40 were highly susceptible (χ2=2·23, P=0·69). This indicated that BB resistance in Ajaya is probably controlled by two additively interacting loci with equal phenotypic effects.
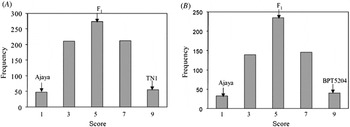
Fig. 1. Frequency distribution of the F2 population derived from Ajaya/TN1(A) and Ajaya/BPT 5204 (B). A segregation ratio of 1:4:6:4:1 with respect to resistance, moderate resistance, moderate susceptibility, susceptibility and high susceptibility was noticed when the F2 plants derived from the crosses Ajaya/TN1(A) and Ajaya/BPT 5204 (B) were screened for BB resistance.
(ii) Analysis with gene-linked markers
In order to test the presence of xa5 and xa13 in Ajaya (as predicted from its pedigree) it was analysed with linked markers RG556 (xa5) and xa13-prom (xa13) along with the NILs of IR24 carrying xa5 (IRBB5), xa13 (IRBB13), the two BB susceptible varieties TN1, BPT 5204 and the parental line BJ1. The CAPS marker RG556 amplified two fragments of sizes 1000 and 1500 bp in all the genotypes. Restriction digestion with the enzyme Dra 1 produced two fragments of sizes 390 and 410 bp in Ajaya, IRBB5 and BJ1 and two fragments of sizes 350 and 450 bp in TN1 and BPT 5204 (Fig. 2(A)). The PCR-based marker xa-13 prom, a functional marker for xa13 amplified the resistance specific allele, a 1·4 kb fragment in IRBB13 and BJ1, whereas Ajaya amplified a 650 bp fragment as in the other susceptible genotypes TN1 and BPT 5204 (Fig. 2(B)).

Fig. 2. Analysis of xa5 (RG556) and xa13 (xa13 prom) linked markers in Ajaya. Ajaya displayed CAPS polymorphism similar to IRBB5, indicating presence of xa5 (2A) when amplified with RG556 and restricted with Dra1. However, it did not show an amplification pattern specific for the resistant allele of xa13, when amplified with the marker xa13 prom (2B). M – Molecular weight marker (100 bp ladder), I5 – IRBB5, I13 – IRBB13, A – Ajaya, BJ – BJ1, T – TN1 and B – BPT 5204.
(iii) Allelism tests
In order to confirm the presence/absence of xa5 and xa13 genes in Ajaya, it was crossed with IRBB5 and IRBB13. The F1s and the F2 population derived from the cross IRBB5/Ajaya was screened with Xoo strain DX-066, which showed resistance reaction with IRBB5 and Ajaya. The F1 was resistant (score 1) and all the 177 F2 plants were uniformly resistant (score 1) without any segregation for susceptibility, indicating that one of the two resistance genes in Ajaya is xa5. The F1 and the F2 population obtained from IRBB13/Ajaya were screened with Xoo strain DX-084, which showed resistance reaction with IRBB13 and Ajaya (score 1). The F1 was highly susceptible (score 9) and out of 326 F2 plants screened, 157 were resistant (score 1) and 169 were highly susceptible.
(iv) Sequence analysis for xa5 candidate gene TFIIAγ
A 244 bp fragment was amplified from the second exon of the xa5 gene candidate gene (i.e. TFIIAγ) encompassing the functional nucleotide polymorphism consisting of the 2 bp substitution GTC/GAG in Ajaya, IRBB5 and two susceptible genotypes IR24 and TN1 and sequenced as explained in Materials and methods. Ajaya possessed the 2 bp polymorphism (TC→AG substitution) specific for xa5 gene, similar to that of IRBB5 at the 17th and 18th nucleotide position, while TN1 and BPT 5204 showed susceptibility-specific TC sequence at the 17th and 18th nucleotides (Fig. 3).

Fig. 3. Sequence alignment of the functional polymorphic region in second exon of xa5. Ajaya possessed the 2 bp polymorphism (TC→AG substitution), which was similar to that of IRBB5 at the 17th and 18th nucleotide position (highlighted in box), while TN1 and BPT 5204 showed susceptible specific TC sequence.
(v) Mapping of the second BB resistance locus in Ajaya
Genetic analysis in the F2 plants derived from the crosses Ajaya/TN1 and Ajaya/BPT 5204 indicated that the action of two loci are involved in conferring BB resistance in Ajaya. Based on the results of analyses with the xa5 linked marker, RG556, allelism test with IRBB 5 and sequence analysis of xa5 candidate gene, the first locus was confirmed to be xa5. The second locus was found to be non-allelic to xa13 (based on linked marker analysis), but linked to it based on allelism test with IRBB13. In order to map the second resistance locus in Ajaya, an analysis was conducted to identify SSR markers polymorphic between Ajaya and TN1. Out of 480 SSR markers tested, 112 (23·3%) were polymorphic and 368 (76·6%) were monomorphic. BSA revealed linkage of the SSR marker RM17745, located on Chr. 5 and RM23535 on chromosome 8 with BB resistance. Out of 22 SSR markers tested in the vicinity of RM 17745 on the short arm of Chr. 5, 10 were polymorphic between Ajaya and TN1. Out of 45 SSR markers tested in the vicinity of RM23535 on the long arm of Chr. 8, nine were polymorphic between Ajaya and TN1. Linkage analysis was performed by genotyping the 800 F2 plants of Ajaya/TN1 with nine SSR markers on Chr. 5, RG556 and nine SSR markers on Chr. 8. The nine SSR markers could be integrated into the linkage map of Chr. 5 and covered a span of 30·5 cM, with mean distance between the markers being 3·38 cM. The nine markers on Chr. 8 spanned a genetic distance of 29·8 cM with a mean distance of 3·31 cM. The marker order on the linkage maps of chromosomes 5 and 8 was consistent with the SSR linkage maps of Temnykh et al. (Reference Temnykh, Park, Cartinhour, Hauck, Lipovich, Cho, Ishii and McCouch2000) and McCouch et al. (Reference McCouch, Teytelman, Xu, Lobos, Clare, Walton, Fu, Maghirang, Li, Xing, Zhang, Kono, Yano, Robert, DeClerck, Schneider, Cartinhour, Ware and Stein2002) .
(vi) QTL analysis
Composite interval mapping (CIM) performed with a set of ten SSR markers on the short arm of chromosome 5 and nine markers on the long arm of Chr. 8 mapped the first resistance locus (tentatively named as qtl BBR 5.1) at a genetic distance of 0·3 cM from RM17751 and 0·5 cM from RM17752 with both the markers flanking the gene at an LOD score of 6·2. RG556 co-segregated with resistance without any recombination indicating that qtlBBR 5.1 is xa5. The second resistance locus (tentatively named as qtl BBR 8.1) was mapped at a genetic distance of 0·2 cM from RM23499 and 0·4 cM from RMAFM1 with both of them flanking the gene (Fig. 4(A)) at an LOD score of 5·7 (Table 2). Marker-trait co-segregation analysis with the marker RM23499 revealed three recombinants in a population of 800 individuals with one recombinant from the moderately susceptible class and two from the susceptible class while with respect to the marker RMAFM1, four recombinants (i.e. two from the moderately susceptible class and two from the susceptible class) were observed. All the 48 highly resistant plants showed amplification of homozygous-resistant parent-specific allele and the 56 highly susceptible plants showed the homozygous susceptible allele of RG 556 and RM23499 without any recombination. The above two loci explained 97·5% of the total variance observed. Since the second resistance locus qBBR8.1 was observed to be non-allelic but linked to xa13 based on allelic testing with IRBB13, it can be considered as a new gene/QTL for BB resistance.

Fig. 4. Genetic linkage map of the genomic region in the vicinity of qtl BBR 5.1 (xa5) and qtl BBR8.1 (xaAj) in the F2 mapping population derived from the cross Ajaya/TN1(A) and Ajaya/BPT 5204 (B). Two major QTLs/genes were identified to be controlling BB resistance in Ajaya. One was xa5 located on Chr. 5 cosegregating with RG556 and the other was xaAj located on Chr. 8 and flanked by the SSR markers RM23499 and RMAFM1.
Table 2. Details of the resistance genes/QTLs in Ajaya identified by composite interval mapping
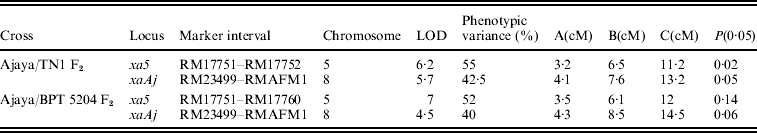
A, additive effect of Ajaya allele; B, lesion length of plants with genotype of Ajaya and C, lesion length of plants with genotype of TN1/BPT 5204.
(vii) Physical mapping
To physically map the locus qBBR8.1, the anchor markers RMAFM1 and RM23499 were used to land on the reference sequence of cv. Nipponbare by BLASTN analysis. The matching sequences showed that there were four PAC clones and three BAC clones, which covered the target gene i.e. OJ1113_A10, P0700D12, OJ1191_A10, P0702E04, P0665C04, OJ1211_G06 and P0623F08. The marker RM23327 was observed to be located on the BAC clone OJ1113_A10 (AP004643), RM23450 on PAC clone P0700D12 (AP004708), RM23468 on BAC clone OJ1191_A10 (AP003888), RMAFM2, RMAFM1 and RM23499 on PAC clone P0702E04 (AP005529), RMAFM5 on PAC clone P0665C04 (AP004464), RM23535 on BAC clone OJ1211_G06 (AP003948) and RM23553 on PAC clone P0623F08 (AP004632). The BAC/PAC clones identified from the IRGSP website were aligned as a contig map via pairwise BLAST analysis. The marker AFM2 aligned at position 60·574 kb on the PAC clone P0702E04, while the flanking markers RMAFM1 and RM23499 aligned at positions 77·895 and 91·330 kb, respectively. Consequently, a physical map covering the qtlBBR8.1 was generated in a 13·5 kb physical interval based on the RGP BAC/PAC contigs (Fig. 5).

Fig. 5. Physical map of xaAj. Nine SSR markers were used in this study. The long horizontal lines indicate the region containing the xaAj. The short horizontal lines represent the BAC/PAC clones of cv. Nipponbare released by IRGSP and assembled by the corresponding markers linked to the xaAj. The numbers at the top above the long horizontal line are the recombination events in the mapping population. The digits between markers are physical distances in kilobase (kb). The vertical and dashed lines denote the relative positions of the corresponding markers.
(viii) Validation of the linked markers in the alternate mapping population
The utility of the linked SSR markers to predict resistance in Ajaya was validated in an alternate F2 population derived from the cross Ajaya/BPT 5204. The xa5 linked markers on chromosome 5 i.e. RG556, RM17720, RM17738, RM17745, RM17751, RM17760, RM17771 and RM17785 were polymorphic while RM17725 and RM17752 were monomorphic between Ajaya and BPT 5204. The markers RM23327, RM23450, RM23468, RMAFM1, RM23499, RMAFM5, RM23535 and RM23553 on chromosome 8 were polymorphic while RMAFM2 was monomorphic between the parents. The marker RG556 displayed perfect co-segregation with resistance. The seven polymorphic SSR markers spanned a genetic distance of 29·2 cM on the linkage map of Chr. 5 with a mean distance of 4·17 cM between the markers, whereas the eight SSR markers on Chr. 8 spanned a genetic distance of 30·55 cM with a mean of 3·81 cM between markers. Composite interval mapping with these markers mapped the first resistance locus (qtl BBR5.1) in the marker interval of RM17751 and RM17760 with an LOD score of 7 and the second resistance locus (qtl BBR8.1/xaAj) in the marker interval of RM23499 and RMAFM1 (Fig. 4(B)) with an LOD score of 4·5 (Table 2). The gene was mapped at a distance of 0·1 cM from RM23499 with a single recombinant (in the moderately resistant class) in a population of 592 individuals while the other flanking marker RMAFM1 showed 0·65 cM distance with five recombinants (one from moderately resistant class, three from moderately susceptible and one from the susceptible class). The 32 highly resistant plants showed homozygous resistant allele and 40 highly susceptible plants showed homozygous susceptible allele with respect to RG556, RM23499 and RMAFM1. The two loci xa5 and xaAj explained 92% of the total phenotypic variance observed. The marker order and the linkage distances between the markers were consistent with that observed in the F2 population derived from Ajaya/TN1. Although there were few recombinants, no recombinants were observed in the highly resistant class with respect to the flanking markers RM23499 and RMAFM1 and so the marker combination of RG556+RM23499 and RMAFM1, could predict complete resistance in Ajaya even in the alternate background of BPT 5204.
(ix) In silico identification of putative candidate genes for qtl BBR8.1
In silico analysis of the intervening genomic region of 13·5 kb flanked by the closest markers RM23499 and RMAFM1 revealed the presence of four putative candidate genes (Table 3), which could be associated with xa5 in imparting complete resistance in Ajaya.
Table 3. Putative candidate genes annotated in the genomic region spanned by xaAj

4. Discussion
Losses due to BB are significantly higher in tropical Asia than in temperate regions because of the prevalence of virulent populations of Xoo. The Indian isolates of Xoo are reported to be more diverse and possess high level of virulence when compared with isolates from other geographic regions (Seshu, Reference Seshu1989). Thus, the resistance genes identified in other geographical regions may not be of significant value for resistance breeding in India. In India, where rice is grown under diverse agro-climatic conditions, a significant diversity in the Xoo pathogen was reported through an analysis of 67 isolates from 18 locations (Yoshitola et al., Reference Yoshitola, Krishnaveni, Reddy and Sonti1997; Shanti et al., Reference Shanti, George, Vera Cruz, Bernardo, Nelson, Leung, Reddy and Sridhar2001). The cultivar, Ajaya possesses high levels of resistance against most of the Indian isolates and has been observed to be durably resistant since its release in 1992 (Laha et al., Reference Laha, Reddy, Krishnaveni, Sundaram, Srinivas Prasad, Ram, Muralidharan and Viraktamath2009).
The choice of durably resistant elite varieties like Ajaya as donors of BB resistance shortens the time required for developing a resistant variety through traditional/marker assisted selection, since it is an already released cultivar with some desirable agronomic features. However, in order to devise a breeding programme for transfer of BB resistance from Ajaya, knowledge about the genetics/mode of inheritance of resistance is necessary. Results from two earlier studies on inheritance of resistance to BB in Ajaya are conflicting. Hence, the present study was devised to study the genetics of BB resistance, tag and map the resistance gene(s)/QTLs in order to facilitate marker assisted introgression. In addition, the study also attempted to identify putative candidates for BB resistance in Ajaya for their possible map-based cloning in future.
Genetic analysis of the F2 population derived from the crosses involving Ajaya with TN1 and BPT 5204 revealed quantitative inheritance of resistance, governed by two loci with equal effects. The quantitative nature of disease resistance involving 2–3 genes has been reported in many species against a variety of pathogens (Leonard, Reference Leonard, Jacobs and Parlevliet1993). The popular IRRI bred varieties IR28, IR26 and the differential cultivar DV85 (Wang et al., Reference Wang, Su, Zhai and Wan2005) were reported to possess quantitative resistance against BB. In the present study, the additive/quantitative interaction between two genes/QTLs with equal effect was discovered and one of these genes/QTLs was confirmed to be xa5 and the same was validated in an alternate mapping population. Based on these results, the durability of resistance in Ajaya can be attributed to its quantitative nature of inheritance of resistance.
Ajaya has a complex pedigree (see Supplementary Figure S1 available at http://journals.cambridge.org/grh) and was originally derived from a cross between IET4141 and CR98-7216. The landrace BJ1, the predicted BB resistance donor of Ajaya has been reported to possess the BB resistance genes xa5 and xa13 (Ogawa et al., Reference Ogawa, Lao, Tabien and Khush1987a). Since genetic analysis of F2 populations in the present study revealed the action of two genes/QTLs involved in conferring resistance in Ajaya, its allelic status was tested with respect to the xa5 linked marker RG556 (Blair & McCouch, Reference Blair and McCouch1997) and a functional marker for xa13, xa13-prom along with its parent BJ1. Iyer & McCouch (Reference Iyer and McCouch2004) reported that the functional polymorphism specific for xa5 is a 2 bp substitution in the 17th and 18th nucleotide positions of exon-2 (i.e. TC→AG) of transcription factor IIAγ (which is the candidate gene for xa5 located on Chr. 5) and this results in the change in a single amino acid (valine→glutamic acid) at the 39th position in the encoded protein. We observed presence of the TC→AG substitution in Ajaya, which conclusively shows that xa5 is indeed one of the genes governing BB resistance in Ajaya.
In an earlier study, Kameswara Rao et al. (Reference Kameswara Rao, Jena and Lakshminarasu2003) reported that a gene non-allelic to xa5 controls BB resistance in Ajaya and mapped it on the long arm of Chr. 5 between the markers RM39 (14·5 cM from the gene) and RM31 (17·7 cM from the gene). xa5 has been conclusively mapped on the short arm of Chr. 5 (Blair & McCouch, Reference Blair and McCouch1997; Iyer & McCouch, Reference Iyer and McCouch2004). In our study, through all the four approaches i.e. marker analysis with the closely linked CAPS marker RG556, allelism test with IRBB5, sequence analysis of the functional polymorphic region of xa5 and linkage analysis in two populations segregating for xa5 we have shown that xa5 is indeed one of the candidate genes conferring resistance in Ajaya, which is in contrast to the observations of Kameswara Rao et al. (Reference Kameswara Rao, Jena and Lakshminarasu2003) in terms of mapping of the recessive resistance gene/QTL on Chr. 5.
The improved resistance of Ajaya to BB as compared with IRBB5, both in terms of the level and spectrum of resistance could be attributed to the additive effect of the second resistance gene/QTL identified in the present study (xaAj) in addition to xa5. Additive/dosage effects have been reported with respect to the major BB resistance genes Xa1 and Xa3 in Java 14 (Kaku, Reference Kaku1997) and between Xa1 and Xa4 in IR20 (Kaku, Reference Kaku1999). The increased level of resistance conferred by more than one gene governing resistance to a single pathogen race has been described as quantitative complementation and has been reported in several studies involving gene-pyramiding studies for BB resistance (Yoshimura et al., Reference Yoshimura, Umehara, Kurata, Nagamuva, Sasaki, Minobe and Iwata1996).
Through QTL analysis in the F2 populations derived from the crosses Ajaya/TN1 and Ajaya/BPT 5204, we fine-mapped the second resistance locus qtl BBR 8.1 which has been designated as xaAj, approximately at a distance of 100 kb from another BB resistance gene xa13. Genetic analysis and QTL analysis in the F2 population revealed equal contribution of resistance by the two loci in Ajaya. The BB differential DV85 was reported to carry two QTLs for BB resistance that were resolved to be xa5 and Xa7 (based on similar map location), with xa5 imparting a larger effect than Xa7 (Wang et al., Reference Wang, Su, Zhai and Wan2005).
When the resistance spectrum of Ajaya was studied with seven hypervirulent isolates of Xoo, three isolates could differentiate Ajaya and BJ1 (which possess xa5 and xa13), two isolates could differentiate Ajaya and IRBB53 (which possesses xa5 and xa13) and five of the seven isolates showed differential disease reaction between Ajaya and IRBB13 (which possesses xa13; Table 1). Based on these observations, the second resistance locus identified in the present study, i.e. xaAj can be considered as novel.
The gene density in the region flanking xa Aj is about one gene every 4·3 kb, against the published predictions of one gene every 9·9 kb (International Rice Genome Sequencing Project, 2005). In the region flanking xa13 (Chu et al., Reference Chu, Fu, Yang, Xu, Li, Sanchez, Park, Bennetzen, Zhang and Wang2006a) and Xa30t (Cheema et al., Reference Cheema, Grewal, Vikal, Sharma, Lore, Das, Bhatia, Mahajan, Gupta, Bharaj and Singh2008), each centimorgan is roughly equivalent to 96 and 60 kb, respectively, whereas in the region flanking xaAj, 1 cM was calculated to be equivalent to 22·5 kb indicating a nearly 12-fold reduction in the ratio of physical/genetic sizes as compared with the average P/G ratio of 260–280 kb/cM estimated by Wu & Tanksley (Reference Wu and Tanksley1993) and Yoshimura et al. (Reference Yoshimura, Umehara, Kurata, Nagamuva, Sasaki, Minobe and Iwata1996) . This reduction is apparently because of enhanced recombination in this region (hotspot for crossing over). In the xa5 region on chromosome 5, each cM is equivalent to about 61 kb (Yang et al., Reference Yang, Sanchez, Khush, Zhu and Huang1998) and 130 kb in the Xa1 region of chromosome 4 (Yoshimura et al., Reference Yoshimura, Umehara, Kurata, Nagamuva, Sasaki, Minobe and Iwata1996).
Plant disease resistance genes frequently occur in the form of tightly linked clusters. Four dominant BB resistance genes Xa3, Xa26t, Xa4 and Xa22 have been mapped to the distal region of chromosome 11L (Yang et al., Reference Yang, Sun, Wang and Zhang2003) and five genes, i.e. Xa1, Xa2, Xa12, Xa14 and Xa30t in a 600 kb region on chromosome 4L (Cheema et al., Reference Cheema, Grewal, Vikal, Sharma, Lore, Das, Bhatia, Mahajan, Gupta, Bharaj and Singh2008). Recessive resistance genes generally do not cluster with other resistance genes in rice (Richter & Ronald, Reference Richter and Ronald2000). However, in the present study xaAj was mapped in a genomic region of approximately 100 kb to xa13. The close linkage of xaAj with xa13 based on CIM analysis was consistent with the results of genetic linkage between the two genes as revealed by allelic testing of Ajaya with IRBB13.
One of the genes annotated through in silico analysis in the genomic region spanned by xaAj encodes a nuclear transport factor which is involved in trafficking of ions and macromolecules between nucleus and cytoplasm. The second gene encodes a hypothetical protein with a conserved Nodulin domain. The candidate gene for xa13 has been identified to be Medicago truncatulata, nodulin gene. Nodulin-related genes are found in several species such as nematodes, insects and animals, although the biochemical functions of their proteins are unknown. Yang et al., (Reference Yang, Sugio and White2006) reported about 17 nodulin-related genes distributed all over the rice genome. The third gene predicted codes for a protein homologous to SUA7, which codes for the general transcription factor TFIIIB or the BRF1 subunit of the transcription factor TFIIB, involved in gene transcription in association with the enzyme RNA Polymerase II. The gene xaAj additively interacts with xa5 in conferring complete resistance to BB in Ajaya based on the results on genetic analysis and CIM analysis. It should be noted that xa5 is a general transcription factor but serves as a resistance-associated protein under BB infection (Iyer & McCouch, Reference Iyer and McCouch2004). The fourth putative candidate gene encodes a peroxidase. Peroxidases are involved in lignin biosynthesis whose accumulation was observed in the resistance mediated by xa5, Xa7 and Xa10 (Reimers & Leach, Reference Reimers and Leach1991). In rice, induction of peroxidases has been correlated with resistance to BB (Young et al., Reference Young, Guo, Guikema, White and Leach1995; Hilaire et al., Reference Hilaire, Young, Willard, McGee, Sweat, Chittoor, Guikema and Leach2001). Based on these, any of the four candidate genes mentioned could be considered as a possible candidate for xaAj and further analysis of these genes through cloning and sequence analysis may reveal their functionality with respect to xaAj. Transcriptional profiling in Ajaya under BB infection revealed the differential expression of defence-related and transcription factor genes, which is in agreement with our results on in silico gene prediction (Kameswara Rao et al., Reference Kameswara Rao, Randeep, Kouji, Junko, Pratibha, Hitoshi and Shoshi2007).
5. Conclusion
The present study has identified two genes/QTLs governing BB resistance in Ajaya, one of which is xa5 and the other linked to, but non-allelic to xa13. The second gene/QTL designated as xaAj was fine mapped on Chr. 8 in a physical interval of 13·5 kb close to (approximately 100 kb) another major BB resistance gene xa13. The gene can be deployed either singly or in combination with other major BB resistance genes xa5, xa13 and Xa21 to obtain broad-spectrum resistance against BB. The tightly linked flanking markers identified for xaAj, i.e. RM23499 and RMAFM1 can be used to track the introgression of the gene into elite backgrounds. The putatively expressed genes, which could be candidates for xaAj, can be validated through map-based cloning. Information obtained from the present study may be valuable in understanding the molecular mechanism(s) behind quantitative resistance to Xanthomonas in rice.
One of the authors (K. Sujatha) gratefully acknowledges the financial assistance in terms of a Junior Research Fellowship and Senior Research Fellowship provided by the Council for Scientific and Industrial Research (CSIR), Government of India for her PhD study. The authors also gratefully acknowledge the financial support provided by the Department of Biotechnology, Government of India and the Indian Council for Agricultural Research, Government of India for carrying out the research study.
6. Supplementary material
The online data are available at http://journals.cambridge.org/GRH












