Introduction
Schistosomiasis is a major threat to over 200 million people globally (Steinmann et al., Reference Steinmann, Keiser, Bos, Tanner and Utzinger2006; Hotez et al., Reference Hotez, Molyneux, Fenwick, Kumaresan, Sachs, Sachs and Savioli2007; Colley et al., Reference Colley, Bustinduy, Secor and King2014; McManus et al., Reference McManus, Dunne, Sacko, Utzinger, Vennervald and Zhou2018). It is caused by the members of the genus Schistosoma, parasitic flatworms, that switch their habitats between a definitive mammalian host and an intermediate snail host (fig. 1). There are three main species that infect humans: S. mansoni; S. japonicum; and S. haematobium. Specific snail hosts are infected by a miracidium, which swims in water using its ciliated plate. Upon infection, the miracidium loses its ciliated plate and transforms into a mother sporocyst. A mother sporocyst reproduces asexually and gives rise to daughter sporocysts that colonize the snail tissue. Daughter sporocysts can produce more daughter sporocysts and undergo embryogenesis, giving rise to hundreds to thousands of cercariae that are released into the water. Cercariae stay alive in water for up to two days, during which they swim forward and reverse using the bifurcated tail to find a mammalian host skin. Upon skin penetration, cercariae lose their tail and the body transforms into schistosomula. Schistosomula migrate across the skin to enter the blood vessel and follow the circulation, first passing through the lung and later arriving at either the hepatic portal vein (S. mansoni and S. japonicum) or the bladder venules (S. haematobium), where they initiate blood feeding. Schistosomes rapidly grow into a male or a female that pair up and reproduce (Basch, Reference Basch1991). The paired worms lay numerous eggs, the majority of which get trapped in host tissues (e.g. liver, spleen and bladder) that attract immune cells, causing inflammation and granuloma formation. The rest of the eggs excreted via urine or faeces hatch in water into miracidia to infect a snail host. Praziquantel has been used for chemotherapy for over several decades in the past, and effectively eliminates adult schistosomes. It activates schistosomes’ transient receptor potential ion channel, causing an influx of calcium that leads to worm paralysis (Park et al., Reference Park, Gunaratne, Chulkov, Moehring, McCusker, Dosa, Chan, Stucky and Marchant2019, Reference Park, Friedrich, Yahya, Rohr, Chulkov, Maillard, Rippmann, Spangenberg and Marchant2021; Le Clec'h et al., Reference Le Clec'h, Chevalier and Mattos2021). However, praziquantel is less active against immature schistosomes, it does not prevent reinfection, and resistance has been documented, calling for new approaches to target these parasites.
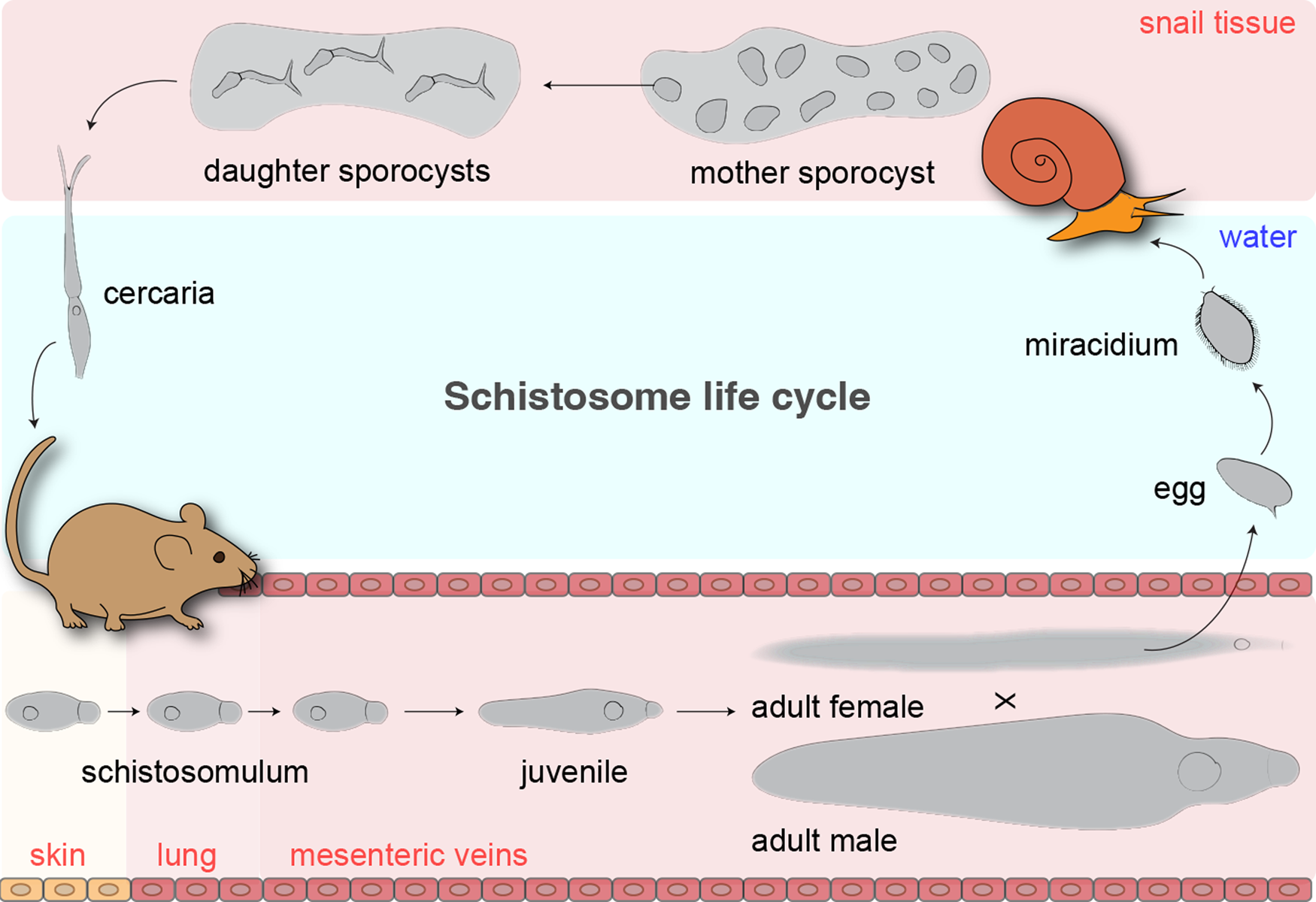
Fig. 1. Life cycle of Schistosoma mansoni.
One of the fascinating aspects of intramammalian stage schistosomes is their ability to thrive inside the host vasculature for decades. In the face of hostile immune components, schistosomes deploy mechanisms from their host–parasite interfaces (i.e. tegument (outer skin) and digestive tract) to evade or neutralize them. For instance, the tegument forms a double lipid bilayer, a structure uniquely found in blood flukes, shortly after the parasites penetrate the skin (McLaren & Hockley, Reference McLaren and Hockley1977). In addition, early studies of the tegument suggest that it can display host glycolipids presumably to evade detection (Goldring et al., Reference Goldring, Clegg, Smithers and Terry1976, Reference Goldring, Sher, Smithers and McLaren1977; Pujol & Cesari, Reference Pujol and Cesari1993). Recently, schistosome stem cells and their deployment throughout the parasite life cycle has been recognized as a primary driver of transmission, homeostasis, reproduction and immune evasion (Collins et al., Reference Collins, Wang, Lambrus, Tharp, Iyer and Newmark2013, Reference Collins, Wendt, Iyer and Newmark2016; Wang et al., Reference Wang, Collins and Newmark2013, Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018; Collins & Collins, Reference Collins and Collins2016; Wendt & Collins, Reference Wendt and Collins2016; Lee et al., Reference Lee, Chong and Newmark2020; Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021; Nanes Sarfati et al., Reference Nanes Sarfati, Li, Tarashansky and Wang2021). In adults, stem cells produce large amounts of tegument cells to constantly replenish cells sloughed off from the host immune attacks (Collins et al., Reference Collins, Wendt, Iyer and Newmark2016; Wendt et al., Reference Wendt, Collins, Pei, Pearson, Bennett, Loukas, Berriman, Grishin and Collins2018). In parallel, stem cells produce cells of the digestive tract for red blood cell processing (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020) and blocking and degrading ingested leucocytes (Lee et al., Reference Lee, Chong and Newmark2020). These studies revealed the identity and the function of tissue-specific transcriptional regulators that are crucial for maintaining and proper functioning of the respective parasite–host interfaces.
Genetic regulators (e.g. transcription factor, RNA-binding protein and nuclear receptor) control the expression of downstream genes required for specific cell types that they are expressed in. Their expressions in undifferentiated stem cells/progenitors are in many cases required for the specification and/or maintenance of differentiated cell types that are necessary for proper tissue function during animal development and homeostasis. Such developmental processes have been extensively studied in free-living planarian flatworms over the past few decades (Reddien, Reference Reddien2013, Reference Reddien2018, Reference Reddien2021; Roberts-Galbraith & Newmark, Reference Roberts-Galbraith and Newmark2015). Planarians and schistosomes are distant cousins, in which they belong to Turbellaria and Trematoda classes, respectively, that are part of the same invertebrate animal phylum, Platyhelminthes (platy: flat; helminth: worm) (Laumer et al., Reference Laumer, Hejnol and Giribet2015; Collins, Reference Collins2017). Planarians have an extraordinary ability to regenerate missing body parts via a population of stem cells called neoblasts (Newmark & Sanchez Alvarado, Reference Newmark and Sanchez Alvarado2002). With the advent of the functional and single cell genomics era, flatworm cellular heterogeneity and underlying intricate mechanisms have begun to unravel. Interestingly, recent studies reveal that despite highly divergent body plans, planarians and schistosomes share an unexpectedly high degree of cell type homology, with notable differences in regulator functions (Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021; Nanes Sarfati et al., Reference Nanes Sarfati, Li, Tarashansky and Wang2021; Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021). Understanding such similarities and differences will help us identify parasite-specific adaptations that can be exploited for intervention. In this review, we provide a high-level overview of cell types and tissues in planarians and schistosomes. In addition, we review major regulators that control cell type specification and maintenance with an emphasis on recent discoveries of schistosome regulators and their functions. We anticipate that such a summary will serve as a valuable resource to the community of schistosome researchers and beyond. Lastly, we convey the significance of studying the basic parasite biology that opens new avenues for translational biology.
Stem cells
Stem cells are traditionally defined as cells that can self-renew and differentiate for a long period of time. Their ability to give rise to different cell types defines their potency. In particular, pluripotency is defined as the ability of stem cells to give rise to all the cell types of the three germ layers (i.e. ectoderm, endoderm and mesoderm) that consist of an entire animal. Planarian flatworms, Schmidtea mediterranea, are free-living flatworms that contain a population of stem cells called neoblasts that fuel animal development, homeostasis and regeneration (Baguna et al., Reference Baguna, Salo and Auladell1989; Newmark & Sanchez Alvarado, Reference Newmark and Sanchez Alvarado2002). They are distributed throughout the mesenchyme with features including round morphology, high nuclear-to-cytoplasm ratio and chromatoid bodies (Coward, Reference Coward1974; Rouhana et al., Reference Rouhana, Weiss, King and Newmark2014). Earlier studies using nucleotide pulse-chase revealed that neoblasts are the only dividing cells in the worm body (Newmark & Sanchez Alvarado, Reference Newmark and Sanchez Alvarado2000). Capitalizing on this aspect, subsequent studies used a combination of an irradiation time course, cell sorting and RNA interference (RNAi) to define mRNAs enriched in neoblasts (Reddien et al., Reference Reddien, Bermange, Murfitt, Jennings and Sanchez Alvarado2005a; Eisenhoffer et al., Reference Eisenhoffer, Kang and Sanchez Alvarado2008), which included proliferative markers, such as histones, pcna (proliferating cell nuclear antigen), mcm (minichromosome maintenance), p53, germ/stem cell-associated RNA-binding proteins including vasa, argonaute, piwi (smedwi), bruno-like and signalling molecules such as fgfr (fibroblast-growth factor receptor). To further demonstrate the pluripotency of neoblasts, a combination of sublethal irradiation, clonal assay and single-cell transplantation was used. Specifically, a single neoblast transplanted into an irradiated planarian host was capable of forming colonies that contained progenies of all three germ layers that subsequently regenerated an entire animal (Wagner et al., Reference Wagner, Wang and Reddien2011). However, not all single neoblasts were capable of whole animal regeneration, indicating a functional and molecular heterogeneity among neoblasts. To investigate the nature of neoblast heterogeneity, single-cell transcriptomic approach was employed, which initially revealed three classes: σ (sigma)-neoblasts; ζ (zeta)-neoblasts; and γ (gamma)-neoblasts (van Wolfswinkel et al., Reference van Wolfswinkel, Wagner and Reddien2014). ζ-neoblasts, and γ-neoblasts expressed transcription factors that govern epidermal (zfp, zinc-finger protein) and intestinal (gata4/5/6, GATA-binding factor) lineage specification, respectively, and therefore were placed under a lower hierarchy than σ-neoblasts that did not express lineage-specific transcription factors. However, using a significantly higher number of cells, multiple subsequent single-cell sequencing studies further deconstructed hierarchical and lineage relationships among the neoblast population (Wurtzel et al., Reference Wurtzel, Cote, Poirier, Satija, Regev and Reddien2015, Reference Wurtzel, Oderberg and Reddien2017; Fincher et al., Reference Fincher, Wurtzel, de Hoog, Kravarik and Reddien2018; Plass et al., Reference Plass, Solana and Wolf2018; Scimone et al., Reference Scimone, Wurtzel, Malecek, Fincher, Oderberg, Kravarik and Reddien2018; Swapna et al., Reference Swapna, Molinaro, Lindsay-Mosher, Pearson and Parkinson2018; Zeng et al., Reference Zeng, Li and Guo2018). In particular, Zeng et al., discovered 12 neoblast subclusters (Nb1 to Nb12) from >7000 cells, which were categorized based on the level of piwi-1 expression (Zeng et al., Reference Zeng, Li and Guo2018). The three classes that were initially identified belonged to one or more subclusters from this analysis. From one of the neoblast subclusters (Nb2) that had high expression of piwi-1, the authors identified a membrane protein TSPAN-1 to generate an antibody against. Using the TSPAN-1 antibody, the authors purified TSPAN-1+ cells and performed a single cell transplantation into an irradiated planarian host. Remarkably, a single TSPAN-1+ cell was able to rescue the entire animal, indicating the pluripotency of Piwi-1high TSPAN-1+ neoblasts. In addition, several other neoblast subclusters expressed lineage-specific transcription factors. Interestingly, another recent study that analysed the cell-cycle status of neoblasts from single cell (sc) RNA-seq revealed that neoblast division could produce one undifferentiated and one fate-specified (lineage-committed) stem cells (Raz et al., Reference Raz, Wurtzel and Reddien2021). These fate-specified neoblasts were capable of dividing and forming colonies that seem to retain pluripotency, suggesting a switch or reversion of their fate through cell division. These data depart from a classical model of hierarchical stem cell fate specification where a fate-specified progenitor undergoes a restricted proliferation and differentiation, to a non-hierarchical single-step model. A recent review discusses the concepts of cellular plasticity that relate to transient cell states rather than cell types (Adler & Sanchez Alvarado, Reference Adler and Sanchez Alvarado2015).
Numerous neoblast genes have been found to be essential using a large-scale RNAi regeneration screen (Reddien et al., Reference Reddien, Bermange, Murfitt, Jennings and Sanchez Alvarado2005a; Wagner et al., Reference Wagner, Ho and Reddien2012). These experiments involve feeding planarians with double-stranded RNA against candidate genes (Sanchez Alvarado & Newmark, Reference Sanchez Alvarado and Newmark1999; Newmark et al., Reference Newmark, Reddien, Cebria and Sanchez Alvarado2003), then cutting the treated worms to identify gene knockdowns that display regeneration defects and/or sublethally irradiating the worms for clonal analysis of stem cell expansion and differentiation. Using such a regime, numerous regulators such as RNA-binding proteins (e.g. smedwi (piwi) and bruli, vasa-1) (Reddien et al., Reference Reddien, Oviedo, Jennings, Jenkin and Sanchez Alvarado2005b; Guo et al., Reference Guo, Peters and Newmark2006), zinc finger proteins (zfp-1) and transcription factors (e.g. p53 and soxP-1) (Wagner et al., Reference Wagner, Ho and Reddien2012) were found to be essential for clonal expansion and differentiation, which led to phenotypes including animal curling, regression, lesions, bloating and lysis. In parallel to such intrinsic regulators, a major driver that determines neoblast fate is the signalling cues it receives from the niche. These cues include molecules associated with conserved signalling pathways such as wingless/integrated (Wnt) and bone morphogenetic protein (BMP) that are highly expressed in muscle cells. Perturbation of many of these signalling components led to altered neoblast cell fate that resulted in defects in homeostasis and regeneration (Witchley et al., Reference Witchley, Mayer, Wagner, Owen and Reddien2013; Reddien, Reference Reddien2018).
Schistosome stem-like cells were classically described as ‘germinal cells’ in sporocyst (Cort et al., Reference Cort, Ameel and Van Der Woude1954; Pan, Reference Pan1980). They have a round cell morphology and a high nuclear-to-cytoplasm ratio akin to planarian neoblasts (Collins et al., Reference Collins, Wang, Lambrus, Tharp, Iyer and Newmark2013). Sporocyst germinal cells extensively divide during the asexual reproduction, giving rise to more germinal cells that undergo embryogenesis to produce cercariae. Classical studies have shown that parasites can be maintained from a monomiracidial infection followed by successive serial transplantation of sporocysts into the molluscan host, indicating that germinal cells include pluripotent cells (Jourdane & Theron, Reference Jourdane and Theron1980). The molecular nature of sporocyst stem cells was first characterized by comparing the transcriptomes of sporocysts at different timepoints, revealing a high degree of conservation to planarian neoblast and germ cells, with an enriched expression of cell cycle genes and RNA-binding proteins (Wang et al., Reference Wang, Collins and Newmark2013). Nucleotide pulse-chase experiments revealed germinal cell heterogeneity, with nanos-2 + and nanos2 − cells proliferating at different rates. Co-expression analysis using fluorescence in situ hybridization (FISH) suggested a potential hierarchical relationship between nanos-2 + and nanos-2 − populations, with nanos-2 + cells representing more undifferentiated germinal cells. To further investigate the heterogeneity, a recent study employed single-cell RNA-seq (scRNA-seq), which revealed three classes of sporocyst germinal cells: κ (kappa, klf +/nanos-2 +); δ (delta, nanos-2 +/fgfrA +); and φ (phi, nanos-2 −/fgfrA +) (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018). Evidence from pulse-chase coupled with FISH placed κ-cells at the top of the hierarchy as embryonic stem cell-like cells, δ-cells that arise from κ-cells to become somatic stem cells, and φ-cells that include lineage-committed progenitors that give rise to differentiated sporocyst tissues. During cercarial embryogenesis, κ-cells and δ-cells are packaged in the body at distinct anatomical locations, with κ-cells located within the germinal cell cluster and ~five δ-cells distributed across the mesenchyme: two sets of cells laterally; and one cell in the mid-body. After a cercaria transforms into a schistosomulum, δ-cells are the first cells to initiate proliferation, between one to two days of entering the mammalian host skin to produce cell types that are needed for the transition (Lee et al., Reference Lee, Chong and Newmark2020). Meanwhile, κ-cells begin to divide a few days later, likely producing cells associated with germline/reproductive development.
The intramammalian-stage schistosomes live for decades inside the blood vessel where hostile immune components are in a constant surveillance, implicating their superb immune evasion capacity. Furthermore, in the face of host immunity, parasites grow and sexually mature for reproduction, laying hundreds of eggs daily, which is the cause of the disease pathology. The cellular basis of such immune evasion and pathology has been proposed to be due to the population of parasite stem cells (Collins & Newmark, Reference Collins and Newmark2013; Wendt & Collins, Reference Wendt and Collins2016). Using irradiation to deplete dividing cells, stem cells were discovered in the mesenchyme that share morphological, molecular and functional features with planarian neoblasts and sporocyst germinal cells (Collins et al., Reference Collins, Wang, Lambrus, Tharp, Iyer and Newmark2013). Subsequent single-cell studies in schistosomula (Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020), juveniles (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018; Tarashansky et al., Reference Tarashansky, Xue, Li, Quake and Wang2019, Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021; Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021) and adults (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020) suggested that stem cell heterogeneity becomes more prominent as parasites mature. One of the new populations that arise in the juvenile stage is ε (epsilon)-cells that express high levels of eled. eled is expressed in both germline and somatic stem cells in juveniles and is also found in a subset of adult stem cells. Importantly, a comparison to planarian neoblasts revealed that ε-cells are most similar to pluripotent neoblasts expressing several transcription factors (e.g. soxP-2, unc4, pax6a and gcm1) (Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021). In addition, other juvenile stem cell subclusters including myoD + μ-cells and μ′-cells shared homologous features with planarian muscle progenitors. A list of major and minor subclasses of schistosome stem/progenitor cells and their potential lineage relationships have recently been reviewed in further detail (Nanes Sarfati et al., Reference Nanes Sarfati, Li, Tarashansky and Wang2021).
Several essential regulators have been identified in schistosome stem cells (table 1). Knockdown of vlg-3 (vasa-like gene), ago2-1 (argonaute) and fgfrA (fibroblast growth factor receptor A) in sporocysts led to a significant reduction in proliferating cells and associated transcripts (Wang et al., Reference Wang, Collins and Newmark2013). Interestingly, while vlg-3 knockdown affects both nanos-2 + and nanos-2 − germinal cells, ago2-1 and fgfrA knockdowns only affected nanos-2-germinal cells, indicating functional heterogeneity of these genes in sporocyst stem cells. Similar to sporocysts, fgfrA knockdown in adults depleted proliferating cells in the soma, which resulted in the downregulation of stem cell genes including h2b and nanos-2 (Collins et al., Reference Collins, Wang, Lambrus, Tharp, Iyer and Newmark2013). Along with fgfrA, a few other genes have been reported to play an important role in juvenile and adult parasites. For instance, cbp1, CBP/p300 family protein, is broadly expressed in stem and early progenitor cells (tsp-2 +), and plays an essential role in repressing cell death-induced over-proliferation of stem cells that lead to organ degeneration (e.g. oesophageal gland) followed by in vivo parasite death (Collins & Collins, Reference Collins and Collins2016). In parallel, knockdown of eled led to a loss of nanos-2 + stem cells in the soma and caused precocious differentiation of germ cells (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018). Together, it is becoming increasingly clear that schistosome stem cells are heterogeneous. Contributing to such heterogeneity are subsets of cell types (or cell states) that express tissue-specific transcription factors that regulate differentiation and/or maintenance of specific cell types/tissues. Several of these studies are highlighted below in the subsequent sections (fig. 2 and table 1).
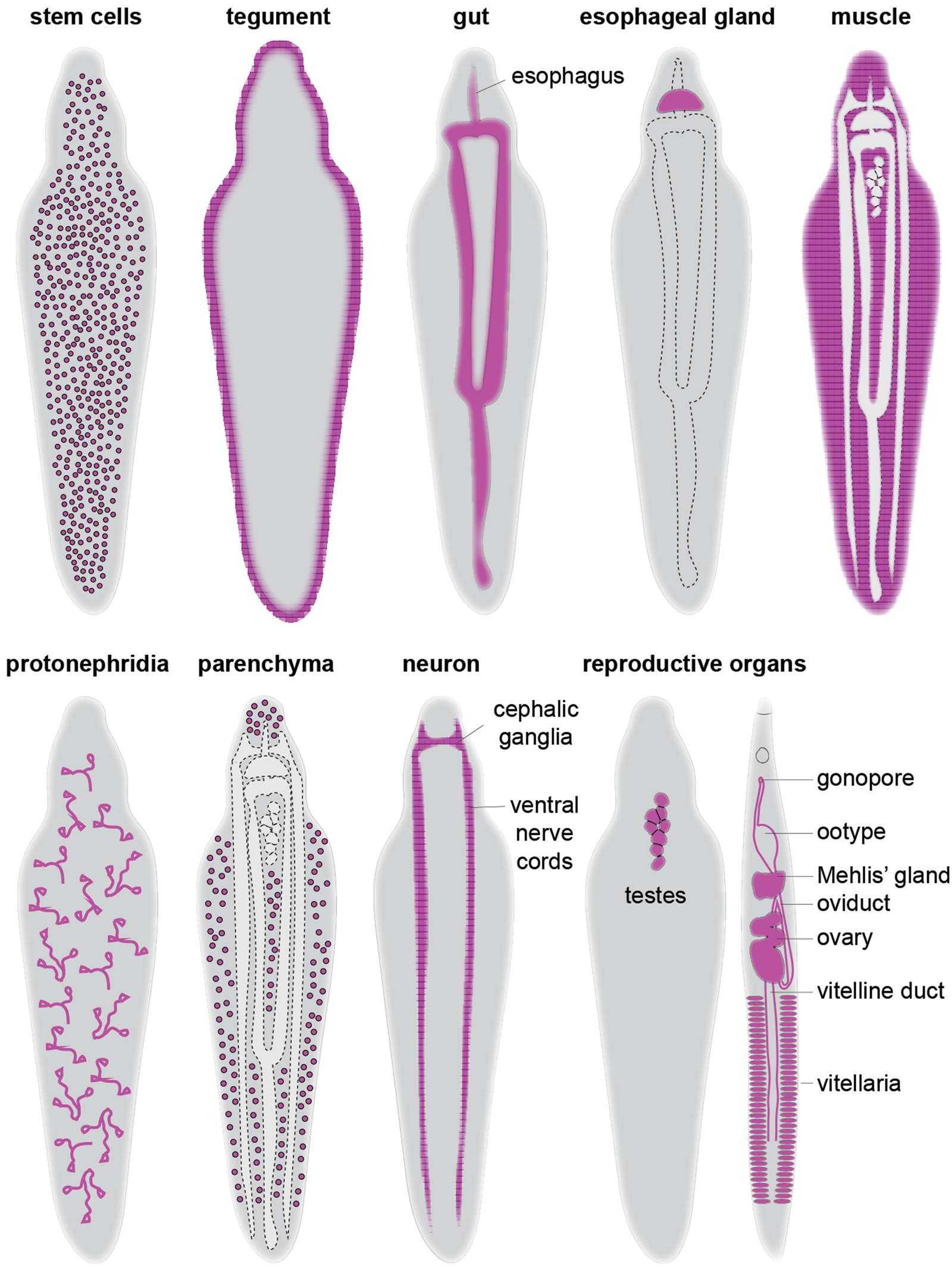
Fig. 2. Schematic overview of schistosome cell types and tissues. For simplicity, a male worm was used to depict the distribution of each cell type except for the reproductive organs, in which both male and female are shown.
Table 1. Genetic regulators of schistosome cell types/tissues and their roles.

Epidermis/tegument
Planarian epidermis has a monostratified layer of cells that form above the basement membrane and make the outer surface of the worm. Epidermal cells consist of differentiated ciliated and non-ciliated cells (Rompolas et al., Reference Rompolas, Patel-King and King2010, Reference Rompolas, Azimzadeh, Marshall and King2013). Early nucleotide pulse-chase studies showed that neoblast continuously give rise to post-mitotic epidermal cells (Newmark & Sanchez Alvarado, Reference Newmark and Sanchez Alvarado2000). Employing irradiation time-course, nucleotide pulse-chase, cell sorting, scRNA-seq and spatiotemporal analysis of epidermal gene expression, multiple studies shed light on cell types and states of epidermal differentiation and identified key genetic regulators (table 2). For instance, zfp-1 and p53, defining markers of ζ-neoblast, play an essential role in maintaining ζ-neoblasts, which are required for the production of epidermal progenitors (Pearson & Sanchez Alvarado, Reference Pearson and Sanchez Alvarado2010; van Wolfswinkel et al., Reference van Wolfswinkel, Wagner and Reddien2014; Cheng et al., Reference Cheng, Tu, Seidel, Robb, Guo and Sanchez Alvarado2018). P53 appears to regulate zfp-1 expression, which in turn regulates the expression of soxP-3 and pax-2/5/8, the two factors that control the transcription of genes required for the secretion of epidermal vesicles (Cheng et al., Reference Cheng, Tu, Seidel, Robb, Guo and Sanchez Alvarado2018). Meanwhile, mex3-1 knockdown led to an expansion of the neoblast pool at the expense of differentiation, resulting in the loss of epidermal progenitors (Zhu et al., Reference Zhu, Hallows, Currie, Xu and Pearson2015). myb-1 appears to be important in activating the transcriptional programme in early epidermal progenitors, and in its absence, the late epidermal differentiation programme was precociously activated (Zhu & Pearson, Reference Zhu and Pearson2018). egr-5, a conserved early growth response family of acetylene (C2H2) zinc finger protein, is not expressed in neoblasts but in early post-mitotic epidermal progenitors (Tu et al., Reference Tu, Cheng, Hanh, Lange, McKinney, Seidel and Sanchez Alvarado2015). Interestingly, knockdown of egr-5 led to misexpression of epidermal differentiation markers resulting in a failure to produce mature epidermis and subsequent loss of epidermal integrity and animal death.
Table 2. Genetic regulators of planarian cell types/tissues and their roles.

Schistosome tegument cells are syncytial and are located beneath the muscle layers. Tegument cells throughout the worm body extend their cytoplasm into the tegument syncytium. The tegument is rapidly rebuilt upon parasites entering the mammalian host, forming a unique double lipid bilayer structure (Wilson & Barnes, Reference Wilson and Barnes1974; McLaren & Hockley, Reference McLaren and Hockley1977; McLaren, Reference McLaren1980; Wilson & Jones, Reference Wilson and Jones2021). This process is thought to be essential for tegument function through the production of proteins and other secreted molecules that enable immune evasion and parasite survival (Wilson & Barnes, Reference Wilson and Barnes1974, Reference Wilson and Barnes1977; Skelly & Wilson, Reference Skelly and Wilson2006). In fact, vaccination of a murine host with TSP-1 and TSP-2, tetraspanin membrane proteins expressed in the tegument, provides significant protection against schistosome infection, with >50% reduction in worm and liver egg burdens (Tran et al., Reference Tran, Pearson and Bethony2006). An extensive review of the composition and function of the tegument and the remaining questions in the field has recently been published (Wilson & Jones, Reference Wilson and Jones2021).
To identify genes underlying tegument formation and maintenance, Collins et al. first used irradiation to deplete adult stem cells/progenitors and compared transcriptomes obtained at different time points after irradiation (Collins et al., Reference Collins, Wendt, Iyer and Newmark2016). From this analysis, the authors discovered that most genes that are downregulated sequentially after the depletion of stem cells were those associated with the tegument. In a subsequent study, the authors used TSP-2 antibody to capture tegument cells from dissociated adult worms that revealed multiple clusters of genes, which allowed them to define the tegument progenitors and mature tegument cells (Wendt et al., Reference Wendt, Collins, Pei, Pearson, Bennett, Loukas, Berriman, Grishin and Collins2018). Further investigation using nucleotide pulse-chase revealed that a large proportion of stem cells produce tegument precursors in both adults (Collins et al., Reference Collins, Wendt, Iyer and Newmark2016) and schistosomula/juveniles (Lee et al., Reference Lee, Chong and Newmark2020). Such continuous replenishment of stem cell-driven tegument cell production likely contributes to the schistosome's ability to evade host immunity.
Investigating the mechanism of tegument cell production, two C2H2 zinc finger proteins, zfp-1 and zfp-1-1, were identified (Wendt et al., Reference Wendt, Collins, Pei, Pearson, Bennett, Loukas, Berriman, Grishin and Collins2018). zfp-1 is highly expressed in a subset of nanos-2 + stem cells and its knockdown led to a slight reduction in mature tegument cells while new gut cell production was significantly impaired, implicating the role of zfp-1 in stem cell differentiation into multiple lineages. Consistent with such findings, zfp-1 was one of the defining markers of juvenile somatic stem cells from single cell analysis (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018). In contrast, zfp-1-1 is not expressed in nanos-2 + stem cells but is highly expressed in early tegument precursors (tsp-2 +). Accordingly, knockdown of zfp-1-1 led to a nearly complete blockage of new tegument cell production, indicating that zfp-1-1 is likely the master regulator of the tegument cell lineage.
Digestive system
Planarian intestine is an epithelial tube that consists of columnar cells that sit above the enteric muscles. Previously, ultrastructural studies identified two major cell types of the planarian gut: phagocytes; and secretory goblet cells (Ishii, Reference Ishii1965). To identify genes enriched in phagocytes, planarians were fed with magnetic dextran particles that were phagocytosed before dissociating the animals into a single cell suspension for cell sorting and transcriptome analysis (Forsthoefel et al., Reference Forsthoefel, James, Escobar, Stary, Vieira, Waters and Newmark2012). From this study, the authors discovered an intestine-enriched homeodomain transcription factor, nkx-2.2, that plays an important role in animal regeneration. nkx-2.2 knockdown led to defects in neoblast proliferation without compromising its ability to differentiate or migrate, eventually resulting in animal death. Interestingly, nkx-2.2 was found to be enriched in γ-neoblasts, along with other conserved endodermal regulators such as hnf4 (hepatocyte nuclear factor 4), gata4/5/6 (GATA-binding factor), and prox-1 (prospero homeobox 1) (van Wolfswinkel et al., Reference van Wolfswinkel, Wagner and Reddien2014). hnf4 and gata4/5/6 knockdown specifically affected nkx-2.2 expressing neoblasts but not the other subclasses (i.e. σ- and ζ-neoblasts), suggesting their role in the endodermal specification of neoblasts.
A recent scRNA-seq revealed a third intestinal cell type besides phagocytes and goblet cells that localizes to the outer epithelial layer with an unknown function (Fincher et al., Reference Fincher, Wurtzel, de Hoog, Kravarik and Reddien2018). Meanwhile, laser-capture microdissection to enrich for intestinal tissues identified additional intestinal genes and cell types (Forsthoefel et al., Reference Forsthoefel, Cejda, Khan and Newmark2020). In this study, the authors were able to determine the identity of three cell types using specific markers, npc2 (Niemann–Pick disease type C2 protein), ctsla (cathepsin La) and slc22a6 (solute carrier family 22 member 6) that labelled goblet cells, phagocytes and basal cells, respectively. Furthermore, npc2 + goblet cells and slc22a6 + basal cells were spatially heterogeneous between the medial (primary) and lateral (secondary and tertiary) branches that contain distinct subsets of cells. Screening for transcriptional regulators, these authors discovered that knockdown of gli-1, a component of hedgehog signalling, perturbed the production of new goblet cells. In contrast, RREB2, ras-responsive element binding protein 2, was required for the maintenance and/or survival of goblet cells. Interestingly, some starved knockdown animals refused to intake food eventually leading to worm lysis and death, suggesting a potential role of goblet cells in regulating hunger.
The planarian digestive system also includes the pharynx, a highly innervated muscular organ that protrudes out to seek and ingest food. It consists of ciliated epithelium, which sits above several muscle layers, neurons and mesenchymal cells. Using chemical amputation-induced pharynx regeneration coupled with comparative transcriptome analysis, a recent study identified forkhead transcription factor, foxA, to be the master regulator of pharyngeal regeneration (Adler et al., Reference Adler, Seidel, McKinney and Sanchez Alvarado2014). foxA is expressed in a subset of neoblasts committed to pharyngeal cell production in both Dugesia japonica (Koinuma et al., Reference Koinuma, Umesono, Watanabe and Agata2000) and S. mediterranea (Adler et al., Reference Adler, Seidel, McKinney and Sanchez Alvarado2014; Scimone et al., Reference Scimone, Kravarik, Lapan and Reddien2014). foxA knockdown animals failed to regenerate all three germ layers comprising the pharynx. Similarly, knockdown of egfr-1, an epidermal growth factor receptor, perturbed neoblast-driven pharynx regeneration (Fraguas et al., Reference Fraguas, Barberan and Cebria2011).
One of the major parasite–host interfaces that plays an important role in immune evasion is the schistosome digestive system. In the worm head, there is a mouth opening (oral sucker) that connects to the oesophagus with surrounding cell masses in the anterior and the posterior halves, the latter of which is a digestive organ called the oesophageal gland. Posterior to the oesophagus are the gut branches that are bifurcated in the anterior half of the worm, which merge into a single branch in the posterior half. Like planarians, schistosomes have a blind gut, and therefore, the waste product of blood feeding, called haemozoin, is regurgitated through the mouth opening (Skelly et al., Reference Skelly, Da'dara, Li, Castro-Borges and Wilson2014). The gut is lined with an epithelial layer, often referred to as gastrodermis (Morris, Reference Morris1968). From scRNA-seq in adult worms, a subset of eled + stem/progenitor cells were found to express hnf4 (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020). Knockdown of hnf4 resulted in an increase in proliferation of eled + stem cells in a place where mature gut tissues would normally form. The lack of new mature gut cells correlated with the downregulation of more than 70% of gut-associated transcripts and defects in digestive capability (e.g. undigested red blood cells in the lumen and reduced protease activity in vitro), ultimately resulting in in vivo parasite death.
The oesophageal gland has been considered as an initial site of blood processing (Li et al., Reference Li, de Castro-Borges, Parker-Manuel, Vance, Demarco, Neves, Evans and Wilson2013) which contains thousands of cell bodies with cytoplasm connected to the oesophageal lumen to release secretory products. Previous ultrastructural studies have identified damaged leucocytes within the lumen, with red blood cells being lysed as they pass through the oesophagus (Dike, Reference Dike1971; Bogitsh & Carter, Reference Bogitsh and Carter1977; Li et al., Reference Li, de Castro-Borges, Parker-Manuel, Vance, Demarco, Neves, Evans and Wilson2013). Following the fate of early stem cells in schistosomula, a recent study discovered that new oesophageal gland cells are produced prior to the initiation of blood feeding, suggestive of an important role that the gland might be playing (Lee et al., Reference Lee, Chong and Newmark2020). Screening through known endodermal regulators, the authors discovered a forkhead transcription factor, foxA, that is enriched in the oesophageal gland and its surrounding stem cells. Knockdown of foxA abolished the oesophageal gland in schistosomula, juveniles and adults. Interestingly, adult parasites lacking the oesophageal gland were unable to block and lyse ingested leucocytes and survive inside the bloodstream, implicating an essential role of the oesophageal gland. Strikingly, parasite death was rescued in immunocompromised mice (e.g. Rag1−/− and μMT−/−), implicating the oesophageal gland as a key organ underlying parasite immune evasion. In addition to foxA, a few other regulators associated with the oesophageal gland have been reported. For instance, cbp1 is broadly expressed in many cell types including stem/progenitor cells and the oesophageal gland. Knockdown of cbp1 led to a degeneration of multiple tissues, including apoptosis of oesophageal gland cells, and resulted in parasite death in vivo (Collins & Collins, Reference Collins and Collins2016). Another example is a nuclear receptor, Ftz-F1, which does not appear to be expressed in the oesophageal gland, that binds to the upstream region of an oesophageal gland-specific gene, meg-8.3 (microexon gene 8.3). Interestingly, Ftz-F1 and meg-8.3 knockdowns led to degeneration of head tissue and loss of parasites’ ability to attach to the culture dish (Romero et al., Reference Romero, Cobb, Collins, Kliewer, Mangelsdorf and Collins2021), highlighting the importance of individual oesophageal gland proteins in parasite homeostasis. Recent biochemical, as well as bulk and single-cell transcriptomic studies have identified several genes enriched in the gland including multiple microexon genes (megs), some of which have been implicated as potential vaccine candidates (Figueiredo et al., Reference Figueiredo, Ricci, de Assis, de Morais, Fonseca and Oliveira2015; Li et al., Reference Li, Xu, Vance, Wang, Lv, van Dam, Cao and Wilson2015; Wilson et al., Reference Wilson, Li and MacDonald2015; Li et al., Reference Li, DeMarco, Neves, James, Newling, Ashton, Cao, Wilson and Castro-Borges2018, Reference Li, Vance, Cartwright, Cao, Wilson and Castro-Borges2020; Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020; Neves et al., Reference Neves, Wilson, Brownridge, Harman, Holman, Beynon, Eyers, DeMarco and Castro-Borges2020; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020).
Germ cells and reproductive organs
S. mediterranea come in two strains: one that reproduces asexually by transverse fission; and the other that reproduces sexually. The sexual strains are simultaneous hermaphrodites carrying male and female germ cells and associated reproductive tissues. Numerous testes are dorsolaterally positioned, while two ovaries are located ventrally at the base of the brain. Yolk glands called vitellaria are ventrally distributed across the worm and produce yolk cells that encapsulate the fertilized embryo (i.e. ectolicithal) before the eggs are laid. The sexual strain of S. mediterranea has served as an excellent model to study germ cell development and regeneration in recent decades (Issigonis & Newmark, Reference Issigonis and Newmark2019). Pioneering work from the Newmark laboratory and others first identified nanos, an RNA-binding protein with a crucial role in germ cell development across metazoan (Seydoux & Braun, Reference Seydoux and Braun2006), to be expressed in presumptive male and female germline stem cells (GSCs), and plays an essential role in germ cell specification and maintenance (Sato et al., Reference Sato, Shibata, Orii, Amikura, Sakurai, Agata, Kobayashi and Watanabe2006; Wang et al., Reference Wang, Zayas, Guo and Newmark2007, Reference Wang, Stary, Wilhelm and Newmark2010). Recently, a Krüppel-like factor associated with pluripotency (Takahashi & Yamanaka, Reference Takahashi and Yamanaka2006), klf4-like (klfl), was found to be expressed in the earliest stage of germ cell development (Issigonis et al., Reference Issigonis, Redkar, Rozario, Khan, Mejia-Sanchez, Lapan, Reddien and Newmark2022; Khan & Newmark, Reference Khan and Newmark2022). klfl is largely co-expressed with nanos in the earliest male and female GSCs as well as in vitellaria and is essential for the specification and maintenance of germ cells and yolk cells (Issigonis et al., Reference Issigonis, Redkar, Rozario, Khan, Mejia-Sanchez, Lapan, Reddien and Newmark2022). In addition to nanos and klfl, multiple factors have been identified to play a role in germ cell development and maintenance. For instance, NF-YB (nuclear factor Y subunit B) is expressed in male GSCs, spermatocytes and spermatids (Wang et al., Reference Wang, Stary, Wilhelm and Newmark2010; Iyer et al., Reference Iyer, Collins and Newmark2016a). Perturbation of NF-YB led to progressive depletion of GSCs, resulting in loss of spermatogonia and degenerated testes over time. boule-1 and boule-2 (deleted in azoospermia family RNA-binding proteins), are expressed in both male and female germ cells and perform distinct roles (Iyer et al., Reference Iyer, Issigonis, Sharma, Extavour and Newmark2016b). boule-1 knockdown led to an expansion of spermatogonia in the expense of meiotic and post-meiotic male germ cells, while boule-2 knockdown resulted in a complete loss of male germ cells, which correlates with increased apoptosis. Meanwhile, boule-1 and boule-2 are not essential for female GSCs’ maintenance but are required to maintain differentiated oocytes. Recent laser-capture microdissection to enrich for reproductive tissues uncovered zfs-1, a zinc finger RNA-binding protein, expressed in female germ cell progenitors, which plays an essential role in regeneration of the entire female germ cell lineage (Khan & Newmark, Reference Khan and Newmark2022).
In parallel to intrinsic factors that regulate germ cell development, there has been a significant advancement in our understanding of extrinsic regulation via local and systemic cues. Within the testes and ovaries, somatic gonadal support cells provide signals for specifying and maintaining germ cells. For instance, dmd-1, a Drosophila melanogaster domain-containing transcription factor, is expressed in male somatic cells and accessory reproductive tissues and is required for the specification and maintenance of nanos + male germ cells and reproductive tissues (Chong et al., Reference Chong, Collins, Brubacher, Zarkower and Newmark2013), revealing the importance of soma–germline communication. Indeed, a recent study highlights the heterogeneity of somatic cell regulation within the female gonad (Khan & Newmark, Reference Khan and Newmark2022). For instance, delta-3 is broadly expressed in ovarian somatic cells whereas foxL, forkhead box transcription factor, is enriched in somatic cells surrounding the late-stage oocytes. In addition to delta-3, other components of notch signalling pathway (e.g. notch-2 and notch-4) were also enriched in female accessory reproductive tissues (e.g. tuba and oviduct). Interestingly, while knockdown of delta-3 and notch-2 led to an expansion and disorganization of klfl + female germ cell progenitors and loss of mature oocytes, foxL knockdown only affected mature oocytes. Beyond the local signalling within the reproductive organs, systemic cues from neural cells also play an important role in germ cell development. For example, pc2, a prohormone convertase, is expressed in the cephalic ganglia, photoreceptors, pharynx, testes and copulatory apparatus, and is required for germ cell differentiation and maintenance (Collins et al., Reference Collins, Hou, Romanova, Lambrus, Miller, Saberi, Sweedler and Newmark2010). PC2 processes several prohormones, including npy-8 (neuropeptide Y-8), which is expressed in subsets of neurons and proximally to testes lobes, and largely phenocopies pc2 (RNAi) animals with degenerated testes lacking mature sperm.
Unlike planarians, schistosomes are dioecious, which is a unique feature among Platyhelminthes. Male and female worms have respective germ cells that produce sperm and egg, as well as associated reproductive tissues (Basch, Reference Basch1991). Male testes are found in the body anterior, and are organized in multiple lobules, each containing undifferentiated germ cells and a mix of differentiating sperm. Female ovary is positioned in the mid-body anterior to where the gut branches merge. In contrast to an unorganized mix of germ cells within a testis lobe, the ovary is well-organized with the most undifferentiated proliferative GSCs positioned anteriorly, while differentiated germ cells are found in the posterior end. Vitellaria are laterally positioned throughout the posterior half of the female and contain differentiating vitellocytes (classically defined as S1, S2, S3 and S4 cells) (Erasmus, Reference Erasmus1975). S1 cells are the most undifferentiated and proliferative, while S4 cells are fully differentiated vitellocytes (Wang & Collins, Reference Wang and Collins2016). Upon fertilization, the embryo is encapsulated by numerous vitellocytes that form an egg that travels through the Mehlis’ gland and ootype and is excreted from the gonopore.
Like GSCs in planarian, nanos-1 expression defines GSCs in schistosomes. Adult scRNA-seq revealed that male and female GSCs share a very similar expression profile which diverges significantly as they differentiate into sperm or egg (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020). In addition, nanos-1 is also expressed in a subset of proliferative (S1) cells in vitellaria (Wang & Collins, Reference Wang and Collins2016). nanos-1 expression arises in juveniles (~2–3 weeks post-infection), which is when germ cells start to develop. Interestingly, eled, a marker initially identified in juvenile stem/germ cells (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018) but is also found in adult stem cells (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020), is expressed earlier than nanos-1 and are later co-expressed, indicating that eled is both the earliest germline and a neoblast marker.
Several functional studies have identified regulators that play an important role in germ cell development and maintenance. eled knockdown in juveniles causes male germ cells to precociously differentiate, leading to an accumulation of meiotic and post-meiotic stage gametes (Wang et al., Reference Wang, Lee, Li, Saberi, Yang, Liu, Zhao and Newmark2018). In contrast, oc-1 (onecut homeobox transcription factor) and boule, expressed in juvenile nanos-1 + GSCs in developing testes and vitellaria, are crucial for promoting male GSC differentiation (Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021). In vitellaria, a nuclear receptor family Vitellogenic Factor 1 (VF1) is expressed in proliferative S1 cells and their immediate progenies and is required for S1 cell differentiation. VF1 knockdown worms failed to produce mature vitellocytes that resulted in decreased egg output without affecting the ovary (Wang et al., Reference Wang, Chen and Collins2019).
One of the unresolved mysteries in schistosomes’ reproductive biology until recently was the mechanism of male-dependent sexual development of females (Basch, Reference Basch1991). The virgin females are significantly shorter than paired females, with premature ovary and vitellaria containing mostly undifferentiated germ cells and yolk cells (Wang et al., Reference Wang, Chen and Collins2019). Only upon physical pairing with a male, a female develops to sexual maturity and starts laying eggs. Screening through major signalling components that are conserved among metazoan, Chen et al. discovered gli-1, a component of hedgehog signalling, to be a key regulator in adult males in activating nrps (non-ribosomal peptide synthetase) (Chen et al., Reference Chen, Wang and Gradinaru2022). gli-1 expression is enriched in a subset of neurons and tegument cells on the ventral side of a male, which is the side that contacts the female. Pairing-induced, gli-1-mediated, nrps activation leads to the production of β-alanine tryptamine (BATT). BATT is secreted from the male worm upon paring, and synthesized BATT was sufficient to induce female sexual development and egg production.
Nervous system
The planarian nervous system consists of two major domains: cephalic ganglia which is the central nervous system; and two ventral nerve cords that run parallel from anterior to posterior of a worm (Agata et al., Reference Agata, Soejima, Kato, Kobayashi, Umesono and Watanabe1998). In accordance with its complexity, scRNA-seq mapped the nervous system with 61 subclusters from >11,000 cells (Fincher et al., Reference Fincher, Wurtzel, de Hoog, Kravarik and Reddien2018). Distinct profiles separate the cell types that make up the brain branches and ventral nerve cords. In addition, many of the clusters included a small subset of cells that can be divided into ciliated and non-ciliated neurons based on the expression of rootletin. Reflective of such great heterogeneity, functional studies over the years have uncovered multiple regulators that are required for maintenance and regeneration of specific neural subsets. For instance, hesl-3, sim and coe are required for proper regeneration of the central nervous system (Cowles et al., Reference Cowles, Brown, Nisperos, Stanley, Pearson and Zayas2013, Reference Cowles, Omuro, Stanley, Quintanilla and Zayas2014). lhx1/5-1 and pitx are required for serotonergic neuron regeneration and maintenance (Currie & Pearson, Reference Currie and Pearson2013; Marz et al., Reference Marz, Seebeck and Bartscherer2013). ap-2 (activating enhancer binding protein) is required for neurons that express TrpA (transient receptor potential cation channel A) (Wenemoser et al., Reference Wenemoser, Lapan, Wilkinson, Bell and Reddien2012; Scimone et al., Reference Scimone, Kravarik, Lapan and Reddien2014), while pax3/7 regulates medial (DBH+) neurons. In parallel, klf4 regulates the regeneration of cintillo + neurons (Scimone et al., Reference Scimone, Kravarik, Lapan and Reddien2014), while soxB1-1 controls the regeneration of sensory neuron subtypes (Ross et al., Reference Ross, Molinaro and Romero2018). Planarians have two dorsolaterally positioned eyespots in the head, each containing numerous photoreceptor neurons. Several factors have been identified as key regulators in regeneration of photoreceptor neurons and pigment cup cells, including six-1/3 (sine oculis), eya (eyes absent), sp6-9 (specificity protein), dlx (distal-less homeobox), soxB1-1 and ovo (ovo/shavenbaby) (Pineda et al., Reference Pineda, Gonzalez, Callaerts, Ikeo, Gehring and Salo2000; Mannini et al., Reference Mannini, Rossi, Deri, Gremigni, Salvetti, Salo and Batistoni2004; Lapan & Reddien, Reference Lapan and Reddien2011, Reference Lapan and Reddien2012). Many of these factors are expressed in neoblasts and early progenitors, highlighting their role in regulating the transcriptional programme governing neural differentiation and maintenance.
The schistosome nervous system also consists of paired cephalic ganglia and ventral nerve cords that run longitudinally through the body. Similar complexity as planarians has been reported, with four and 30 identifiable subclusters in schistosomula and adults that have enriched expression of neuroendocrine protein 7B2 (Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020). In schistosomula, Sm-kk7, ndf (neurogenic differentiation factor), and gnai expressions defined the subclusters while in adults, the sub-clusters belonged to distinct structures within the nervous system, including the cephalic ganglia and peripheral neurons, often with patterns of asymmetry. In addition, like planarians, multiple sub-clusters belonged to either ciliated or non-ciliated neurons. While several transcriptional regulators can be identified from scRNA-seq, including ndf, their function in the development or maintenance of specific neural subtypes remains unknown. The only known example is gli-1, which, as described earlier, is induced in the subsets of ventrally located neurons in males upon pairing with a female, and induces nrps expression to release BATT, promoting female sexual development.
Muscle cells
Coordination of muscles are important for animal movement and feeding. Body wall muscle under the epidermis consists of fibres that are circular, longitudinal and diagonal. Intestinal branches and pharynx also have surrounding musculatures. Recent efforts by Scimone et al., using scRNA-seq revealed expression profiles of DVM (dorsal-ventral muscle), IM (intestinal muscle) and pharynx muscle (Scimone et al., Reference Scimone, Cote and Reddien2017, Reference Scimone, Wurtzel, Malecek, Fincher, Oderberg, Kravarik and Reddien2018). From this study, several regulators that are crucial for muscle regeneration were discovered. For instance, foxF-1 specifies all non-body wall muscles. nk4 and gata4/5/6-2 specify lateral and medial DVM, respectively. In addition, myoD and nkx1-1 were shown to be crucial for specifying body wall longitudinal and circular fibres.
Position control genes that provide signals for proper morphogenesis and development, are expressed in muscle cells in a unique pattern throughout the body (Witchley et al., Reference Witchley, Mayer, Wagner, Owen and Reddien2013; Reddien, Reference Reddien2018). The most well-known are Wnt and BMP, which are crucial for establishing anterior to posterior and dorsal to ventral polarity axes, respectively (Reddien, Reference Reddien2011). For instance, in the absence of β-catenin, a downstream effector of Wnt signalling cascade that stimulates Wnt-specific genes, planarians fail to form a posterior polarity that results in two-headed planarians (Gurley et al., Reference Gurley, Rink and Sanchez Alvarado2008; Petersen & Reddien, Reference Petersen and Reddien2008). Conversely, notum, an inhibitor of Wnt signalling, is expressed in a subset of muscle cells in the head, and is essential for setting up the anterior polarity (Petersen & Reddien, Reference Petersen and Reddien2011). A recent article takes a deep dive into the complex signalling cues that regulate neoblast-driven planarian development and regeneration (Reddien, Reference Reddien2018).
Schistosome muscles in schistosomula, juveniles, and adults display heterogeneity with three to eight identifiable subclusters (Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020; Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020). Adding to their heterogeneity is sex-specific muscle clusters that are only found in females surrounding the ovary. Like planarian muscles, signalling molecules such as Wnt and BMP are highly enriched in specific parts of body muscles (e.g. wnt-2 in the anterior of the worm head). These signals likely play a crucial role in providing instructional cues to stem cells during parasite development and homeostasis. Known master regulators such as myoD and forkhead box protein orthologous to planarian foxF are expressed in a cluster of muscle cells (Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021). However, no functional role of any of the regulators have been reported to date.
Excretory system
Excretory function is essential for maintaining osmotic balance required for homeostasis and survival. The planarian excretory system is organized in a filtration unit called protonephridia, which is distributed throughout the entire worm body. Each protonephridia unit contains a flame cell (ciliated terminal cell), proximal and distal epithelial tubules, collecting duct and tubule-associated cells (McKanna, Reference McKanna1968a, Reference McKannab; Ishii, Reference Ishii1980; Fincher et al., Reference Fincher, Wurtzel, de Hoog, Kravarik and Reddien2018). A comparison of structure and function between protonephridia and vertebrate nephron reveals a striking homology, with distinct functional segments of proximal and distal tubules and collecting ducts that share expression of conserved solute carrier proteins with known functions in transportation of specific ions (Thi-Kim Vu et al., Reference Thi-Kim Vu, Rink, McKinney, McClain, Lakshmanaperumal, Alexander and Sanchez Alvarado2015). Protonephridia completely regenerate within six days, with flame cells arising between one and two days, followed by proximal tubule cells between two and three, and distal tubule cells between three and four days (Rink et al., Reference Rink, Vu and Sanchez Alvarado2011). Several regulators play an important role in producing different cell types. For instance, egfr-5 (epidermal growth factor receptor homolog) is expressed in flame cells and its knockdown led to a loss of flame cells that resulted in branching defects in tubules and oedema formation in homeostatic and regenerating worms (Rink et al., Reference Rink, Vu and Sanchez Alvarado2011). A separate study discovered that hunchback is broadly expressed in the excretory tissue while POU2/3 (Pit-1/Oct1-/2, Unc-86) and six1/2-2 (sine oculus) are enriched in tubule and tubule-associated cells, respectively (Scimone et al., Reference Scimone, Srivastava, Bell and Reddien2011). Knockdown of each of the genes led to a reduction in maintenance and regeneration of flame cells and tubule/tubule-associated cells, which resulted in bloating, blistering and worm lysis. Transcriptomic analysis of knockdown animals revealed a significant downregulation of conserved genes associated with ion transport, protein clearance and acid–base balance-associated enzymes. Similarly, eya (eyes absent) knockdown animals displayed a reduction in tubule and tubule-associated cells in blastema while flame cells were also lost in intact eya (RNAi) animals. Meanwhile, sall (Drosophila splat) knockdown resulted in a decreased number of tubule cells and an increased number of tubule-associated cells, suggesting its role in directing tubule cell fate.
In schistosomes, protonephridia are present in all stages. Intramammalian stage protonephridia begin to develop during cercarial embryogenesis inside the snail tissue. While a previous ultrastructural study suggested that cercaria has six pairs of flame cells (Dorsey et al., Reference Dorsey, Cousin, Lewis and Stirewalt2002), immunostaining with tubulin antibodies revealed five pairs of flame cells distributed in the body: a pair in the anterior body; a pair in the mid-body; two pairs in the dorsal and ventral side of the posterior body; and a pair in the anterior tail (Collins et al., Reference Collins, King, Cogswell, Williams and Newmark2011). As parasites mature, protonephridia number increases and are widely distributed throughout the body, indicating stem cell-driven protonephridia production during parasite development. scRNA-seq identified one cluster of ciliated flame cells that are enriched with sialidase expression (Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020). Interestingly, known regulators such as POU2/3 and egfr-5 are also enriched in this cluster, indicating their potential role in production of protonephridia cells. The exact function of such regulators and the mechanism of protonephridia development in schistosomes remains unexplored.
Other cell types
Classically, planarian parenchyma included several cell types including neoblasts, gland cells and phagocytic cells that are not part of any specific organs but fill up the parenchymal space (Pedersen, Reference Pedersen1959, Reference Pedersen1961). Accordingly, single-cell studies identified eight subclusters of smedwi-1 + precursors and 13 differentiated cell sub-clusters (Fincher et al., Reference Fincher, Wurtzel, de Hoog, Kravarik and Reddien2018). These subclusters represented spatially and functionally distinct cell types, including dorsal, ventral and lateral (marginal adhesive) glands and large cells around the pharynx. Interestingly, nkx-6, a NK homeobox expressed in dd_515 + parenchymal cells, was required to maintain dd_515 + cells. Another interesting group of cells was identified based on the expression of cathepsin. These cells have specialized morphology and express proteases, including Cathepsins that are found in lysosomes. They are distributed across the worm in different tissues, including the brain, around the gut branches and protonephridia, subepidermal and parenchyma space, and pharynx. These cathepsin + cells also included glial subclusters, pigment cells and precursors to these cells. A recent study has shown that fluorescent proteins and bacteria can be taken up by these cells, indicative of their phagocytic nature (Scimone et al., Reference Scimone, Wurtzel, Malecek, Fincher, Oderberg, Kravarik and Reddien2018). Interestingly, foxF-1, which is expressed in both non-body wall muscle cells and cathepsin + cells, is required to maintain cathepsin + cells. Moreover, foxF-1 knockdown worms showed a significant reduction in fluorescent bacteria uptake in cathepsin + cells, which correlates with the loss of phagocytic cells. In schistosomes, two clusters of parenchymal cells were identified from schistosomula and adults (Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020), while two cathepsin + cells and a single parenchymal cell-type were discovered from juvenile (Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021) scRNA-seq. These cells display long processes that occupy the parenchymal space and express proteases such as cathepsin B. Interestingly, foxF-1 is enriched in both parenchymal/cathepsin+ cell clusters (Diaz Soria et al., Reference Diaz Soria, Lee and Chong2020; Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020; Tarashansky et al., Reference Tarashansky, Musser, Khariton, Li, Arendt, Quake and Wang2021) suggesting its putative role in the production and/or maintenance of these cells. However, the mechanism of parenchymal cell development and the role of parenchymal cells in parasite physiology and host–parasite interaction remains unknown.
Conclusions and perspectives
As evidenced in this review, stem cells deploy developmental mechanisms through tissue-specific transcriptional regulators that determine their fate. These mechanisms ensure the proper production of specific cell types needed for tissue development and/or maintenance. Failure to produce or maintain such cell types could lead to an organ failure that renders parasites unfit for survival. Building upon the biology of planarians, several recent studies have begun to uncover tissue-specific regulatory programmes, including tegument cell production by zfp-1-1 (Wendt et al., Reference Wendt, Collins, Pei, Pearson, Bennett, Loukas, Berriman, Grishin and Collins2018), gut cell production by hnf4 (Wendt et al., Reference Wendt, Zhao, Chen, Liu, O'Donoghue, Caffrey, Reese and Collins2020), oesophageal gland cell production by foxA (Lee et al., Reference Lee, Chong and Newmark2020), germ cell differentiation by oc-1 and boule (Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021) and vitellocyte production by VF1 (Wang et al., Reference Wang, Chen and Collins2019). Such discoveries have significant implications in several aspects. First, these studies enhance our understanding of the mechanism of cell-type specification and maintenance. RNAi followed by nucleotide pulse-chase coupled with in situ hybridization revealed how stem cell proliferation and/or differentiation towards a specific lineage is affected in the context of parasite development or homeostasis. While terminally differentiated tissues between planarians and schistosomes are highly divergent in their structure and function (e.g. planarian intestine vs. schistosome gut, pharynx vs. oesophageal gland and epidermis vs. tegument), the identity of endodermal (i.e. hnf4 and foxA) and ectodermal (i.e. zfp-1-1) regulators that govern respective cell type specification are highly conserved. In contrast, despite the identity of such regulators being conserved, the mechanisms of regulator function are often divergent. For instance, although the expression of oc-1 and boule in nanos-1 + germ cells are conserved, their functions are essentially the opposite, where they promote germ cell differentiation in planarians but repress it in schistosomes (Li et al., Reference Li, Nanes Sarfati, Xue, Yu, Tarashansky, Quake and Wang2021). As such, exploiting the shared identity of conserved regulators, we can discover parasite-specific adaptations that can lead to additional intervention strategies. Second, transcriptome analysis of regulator RNAi allowed the identification of specific genes that are dysregulated. Such datasets, together with single-cell transcriptomes, were instrumental in determining the molecular profile of the perturbed cell types/tissues. In addition, tools to enrich for specific tissues (e.g. cell sorting and laser-capture microdissection) allowed a deeper dive into cellular heterogeneity. For instance, TSP-2 antibody-mediated cell dissociation followed by transcriptome analysis identified multiple tegumental cell clusters that are nicely corroborated in subsequent scRNA-seq studies. Third, a combination of in vitro (e.g. movement, attachment, feeding, pairing and death) and in vivo assays (e.g. surgical transplantation) enabled the discovery of how the cell type function relates to parasite homeostasis and survival. For example, the effects of hnf4 and foxA knockdowns (that specifically affect the production of gut and oesophageal gland cells, respectively) were first tested using in vitro feeding of either red or white blood cells. Subsequent in vivo transplantation revealed that neither parasites could survive in the host bloodstream, revealing the essential function of gut and oesophageal gland cells. Fourth, as seen in several examples such as transcriptional regulation of nrps by Gli-1 and meg-8.3 by Ftz-F1, functional screen using RNAi of downstream genes identified from gli-1 and ftz-F1 (RNAi) RNA-seq discovered essential factors that phenocopy the regulator knockdown phenotypes (i.e. failed female sexual development in gli-1/nrps knockdowns and degenerated oesophageal gland and attachment failure in ftz-F1/meg-8.3 knockdowns), bringing mechanistic insights. Lastly, RNAi screen led to the identification of specific factors that are essential for parasite physiology and/or survival (e.g. BATT, MEG-8.3). Such molecules may have translational value as potential therapeutic or intervention targets. Accordingly, a recent study employing a large-scale RNAi screening was proven to be highly successful in identifying druggable targets (Wang et al., Reference Wang, Paz and Padalino2020).
These implications logically lead us to ask several important questions regarding future research directions. First, what are the regulators governing other cell types that are likely to play a vital role in parasite physiology? For instance, like planarians, schistosome muscles, parenchyma, protonephridia and neurons all express several conserved regulators such as myoD, foxF-1, egfr and ndf, just to name a few. Whether these factors indeed regulate the production and/or maintenance of respective cell types, and how such mechanisms might differ from those of planarians remains to be determined. An RNAi screen of candidate regulators coupled with a phenotypic characterization of parasite behaviour (e.g. motility, attachment, migration and feeding) and physiology (e.g. bloating and lysis) would be a viable entry point to answering this question. In addition, employing newly emerging single-cell genomic tools such as spatial transcriptomics would significantly enhance our understanding of the cell type heterogeneity and identify additional regulators (Moffitt et al., Reference Moffitt, Lundberg and Heyn2022). Second, what are the regulators that govern parasite development in other life cycle stages? Most of the currently identified regulators, other than sporocyst germinal cell regulators, are specific to intramammalian stage schistosomes. Understanding the regulatory mechanisms of embryogenesis of miracidia and cercariae, and sporocyst development, could collectively contribute to our understanding of parasite biology and illuminate multiple facets of intervention strategies to disrupt parasite transmission. A combination of known conserved regulators of planarian embryogenesis (Davies et al., Reference Davies, Lei and Seidel2017), existing functional genomics toolkits highlighted above, and recent advancements of transgenic tools in schistosomes (Ittiprasert et al., Reference Ittiprasert, Mann and Karinshak2019; You et al., Reference You, Mayer and Johnston2021; Hulme et al., Reference Hulme, Geyer and Forde-Thomas2022), would serve as an excellent starting point in tackling this question. Lastly, how are such regulatory mechanisms deployed in other parasitic flatworms? Other parasites within the Trematoda class include lung (e.g. Paragonimus) and liver (e.g. Clonorchis, Opisthorchis and Fasciola) flukes. In addition, other classes within the Platyhelminthes phylum include parasites that belong to Monogenea and Cestoda. Monogenea are typically ectoparasites that live on aquatic vertebrates while Cestoda include tapeworms (e.g. Taenia, Echinococcus and Hymenolepis) that have segmented body plans (i.e. proglottids). Interestingly, recent studies have discovered neoblast-like stem cells in liver flukes (McCusker et al., Reference McCusker, McVeigh and Rathinasamy2016) and tapeworms (Rozario et al., Reference Rozario, Quinn, Wang, Davis and Newmark2019), and have begun to explore the molecular mechanisms governing their development and/or regeneration. Collectively, these studies will bring insights to developing new targeting strategies and contribute to the fight against countless devastating parasitic diseases.
Acknowledgements
We thank Bo Wang, Rosa Mejia Sanchez and the members of the Lee Laboratory for their critical input in revising the manuscript.
Financial support
This work was supported by the Dean's startup fund and the Rising STAR award made available to J.L. by the University of Texas Systems.
Conflict of interest declaration
None.
Ethical standards
Not applicable.








