INTRODUCTION
Campylobacter is a Gram-negative, thermophilic, obligate microaerophilic bacterium, with Campylobacter jejuni accounting for 99% of the reported Campylobacter-associated human illness in North America and the UK [Reference Tauxe, Nachamkin, Blaser and Tomkins1]. C. jejuni colonizes the intestinal mucosa of most warm-blooded hosts, including domestic animals and humans, and causes sporadic and epidemic outbreaks of gastroenteritis, particularly in immunocompromised individuals, such as patients with cancer, AIDS or diabetes, as well as very young (especially those aged <7 years) and older people [Reference Park2]. In addition, increasing evidence reveals that some serotypes of C. jejuni are associated with the development of Guillain–Barre syndrome (GBS) in humans [Reference Godschalk3]. C. jejuni is becoming the most common cause of gastroenteritis in developed countries and raises major public health concerns worldwide [Reference Nachamkin4, Reference Skirrow and Blaser5]. Animals such as chicken and cattle may act as asymptomatic reservoirs, by shedding C. jejuni in their stools and contaminating animal food products and surface water during slaughter and carcass dressing. Various reports from Europe and the USA have demonstrated C. jejuni contamination in bovine faecal samples [Reference Dodson and LeJeune6]. Direct or indirect cross-contamination from stools to raw milk may occur in the process of milking, which ultimately causes foodborne outbreaks of C. jejuni after consumption of raw or inadequately pasteurized cows' milk [Reference Jayarao7]. In poultry, contamination with C. jejuni is commensal and is found at the highest level in the mucosal crypts of the caecum and, to a lesser extent, in the small intestine [Reference Hood, Pearson and Shahamat8]. Cross-contamination of commonly consumed poultry products is one possible link between poultry and human campylobacteriosis [Reference Hood, Pearson and Shahamat8]. To explore the role of poultry and dairy cows as infectious sources of C. jejuni in sporadic infections in China, we investigated faecal samples from poultry, dairy cows and diarrhoea patients by multiplex PCR, and classified the isolates by fla-PCR–RFLP.
METHODS
Sampling of human patients
During the period July 2005–December 2006, stool specimens from patients with diarrhoea were collected from three different hospitals (A, B and C) at Yangzhou, Jiangsu Province, China. Details of the samples are given in Table 1.
Table 1. Samples from diarrhoea patients
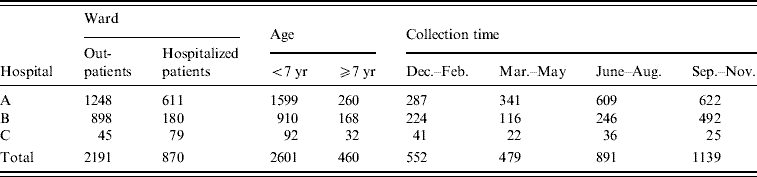
During the period July 2005–December 2006, 3061 stool specimens from patients with diarrhoea were collected from three different hospitals at Yangzhou (hospitals A, B and C). Samples sorted according to ward, patients' age and collection time.
Sampling on cattle farms
A total of 618 rectal samples from Holstein (Bos taurus) milk cows, 368 rectal samples from Holstein heifers, and 545 soil and water samples were collected from 10 cattle farms in Jiangsu Province between March 2005 and June 2006. Soil samples were collected by scraping the surface soil on animal farms with wet swabs, and water samples by direct aspirating or swabs of surface water.
Sampling on poultry farms
A total of 2609 faecal samples were collected from adult chickens aged ⩾6 months in 30 breeder or commercial poultry flocks in the middle, southern and eastern parts of China. These flocks included Chinese native breeds Luo-man, Ai-jiao, Pu-dong, Hai-lan, Xiao Shao-xing, Xin Yang-he, Xin-hui, Qin-ma, Non-da, Huang-ma, Guang-xi, Wu-gu, Hai-hua, Ma-hua, Ma-huang and Tu-ji. In addition, another 532 faecal samples from free-range chickens and 217 soil and water samples were collected from chicken farms using to the above procedures.
Sample preparation
Samples were taken by wiping out the cloacal or rectal contents with sterile swabs and stabbing the swabs into Cary–Blair transport medium, and transporting them to the laboratory within 24 h. All swab samples were transferred into 500 μl sterile PBS, left at room temperature for 20 min, and vortexed every 5 min to release the bacteria from the swabs. Five hundred microlitres of the suspension was added to 9·5 ml Preston Campylobacter Selective Enrichment Broth (Preston Broth supplemented with Preston Campylobacter Selective Supplement SR117, Campylobacter Growth Supplement SR84 and 5% bovine serum albumin (Oxoid, UK), and incubated under microaerobic conditions of 83% N2, 10% CO2, and 7% O2 at 42°C for 36 h.
DNA extraction
Bacterial cells were harvested by centrifugation and resuspended in a 1·5-ml microtube that contained 1 ml sterile double-distilled water. The suspension was adjusted to an optical density of 1·0 at a wavelength of 610 nm using a Hitachi U-1100 spectrophotometer (Eppendorf, Germany). A 400-μl aliquot of adjusted cell suspension was transferred into another microtube containing 0·8 μl lysozyme (50 mg/ml stock) and incubated at 37°C for 20 min. One microlitre of 10 mg/ml RNase, 50 μl 10% SDS was added to the suspension and incubated at 37°C for 30 min. One microlitre of 20 mg/ml proteinase K was added and incubated at 37°C for 90 min. An equal volume of phenol was added to the suspension and was shaken vigorously by hand for 5 min, and then centrifuged at 12 000 rpm for 10 min. The upper phase was transferred into another microtube and genomic DNA was precipitated with 99% ethanol and 0·3 m sodium acetate at −20°C for 30 min. The mixture was then centrifuged at 12 000 rpm for 10 min and the upper phase was discarded. The pellet was washed with 70% ethanol. The DNA pellet was resuspended in 100 μl sterile distilled water and stored at −20°C as a template for the PCR.
PCR identification
Multiplex PCR (mPCR) was carried out according to the method described by Denis et al. [Reference Denis9].
Three sets of primers that targeted 16S rRNA:
forward: 5′-ATCTAATGGCTTAACCATTAAAC-3′;
reverse: 5′-GGACGGTAACTAGTTTAGTATT-3′,
mapA gene:
forward: 5′-CTATTTTATTTTTGAGTGCTTGTG-3′;
reverse: 5′-GCTTTATTTGCCATTTGTTTTATTA-3′,
and ceuE gene:
forward: 5′-AATTGAAAAATTGCTCCAACTATG-3′;
reverse: 5′-TGATTTTATTATTTGTAGCAGCG-3′
were used for the identification of Campylobacter genus, C. jejuni and C. coli, respectively. In a typical reaction, 20 μl PCR system contained 2 μl 10× PCR buffer, 3 pm of each dNTP, 20 pm 16 S1 and 16 S2 primers, 37·5 pm mapA1, mapA2, ceuE1 and ceuE2 primers, 2 μl DNA, 0·2 μl Taq polymerase (Takara Diagnostics, China). The reactions were performed with a thermal cycler (Bio-Rad, USA) using the following temperature-cycling parameters: initial denaturation at 95°C for 10 min, followed by 35 cycles of amplification. Each cycle consisted of 30 s at 95°C, 90 s at 59°C, 1 min at 72°C, and a final extension step of 10 min at 72°C. Amplification generated 857-, 589- and 462-bp DNA fragments that corresponded to the genus Campylobacter, C. jejuni and C. coli, respectively. PCR products (8 μl) were analysed on a 1% agarose gel that contained 0·3 μg/ml ethidium bromide in 0·5× TAE buffer and viewed under UV light. A 100-bp DNA ladder (Promega, USA) was used as a molecular size standard.
Isolation of strains
C. jejuni-positive samples by mPCR were cultured and streaked onto CCDA agar plate and incubated at 42°C under microaerobic conditions of 83% N2, 10% CO2 and 7% O2 for 48 h. After incubation, light grey colonies 1 mm in diameter were subcultured and Gram-stained. The germ colonies from the subculture were then assayed for motility, production of oxidase and catalase, and other biochemical tests [Reference Denis9]. Finally, these isolates were assayed by hippurate hydrolysis.
PCR–RFLP assay
PCR–RFLP analysis was performed as described previously [Reference Nachamkin, Bohachick and Patton10], with some modifications. Briefly, the oligonucleotide primers for flaA gene amplification have been described by Nachamkin et al. – forward: 5′-GGATTCGTATTAACACAAATGGTGC-3′; reverse: 5′-CTGTAGTAATCTTAAAACATTTG-3′ [Reference Nachamkin, Bohachick and Patton10]. The PCR amplification was performed in a Biometra gradient thermal cycler (Bio-Rad), in a total volume of 100 μl, which contained 5 μl DNA, 10 μl 10× PCR buffer, 8 μl each primer at 2·5 μm, 8 μl 2·5 mm dNTP mix, 2·5 U Taq polymerase (Takara Diagnostics). Thermocycling conditions included an initial hold at 94°C for 1 min, followed by 35 cycles of 92°C for 30 s, 55°C for 90 s and 72°C for 105 s. A final extension step of 72°C for 10 min was followed by holding at 4°C. An 8-μl aliquot of each PCR product was run on a 1% agarose gel to confirm successful amplification of the flaA gene. The remaining 80 μl of each PCR product was purified by using 99% ethanol and 0·3 m sodium acetate, and resuspended in 25 μl sterile distilled water. The purified PCR product was digested with DdeI restriction enzyme (10 U/μl; Roche Diagnostics, Germany) in a final reaction volume of 20 μl, and kept at 37°C overnight. The DdeI-digested products were detected on 2% agarose gels in 0·5× TAE buffer and photographed under UV light.
To evaluate the differentiation capability of PCR–RFLP, 10 isolates of C. jejuni were randomly selected for flaA gene amplification. To confirm the reproducibility of this molecular method, the experiments were performed again 2 weeks later.
Statistical analysis
Significance of the clinical study was determined using Fisher's exact test. P<0·05 was regarded as significant, P<0·01 as extremely significant. Positive and negative predictive values were calculated by the χ 2 test.
RESULTS
PCR detection of C. jejuni
PCR products from reference strains of C. jejuni (ATCC33560), C. coli and Campylobacter spp. resulted in bands of 589 and 857 bp, 462 and 857 bp, and 857 bp, respectively. No amplification product was observed for Escherichia coli as a non-campylobacter control. Distinct PCR products were observed for all of the isolates.
Prevalence of C. jejuni in diarrhoea patients
Out of 3061 diarrhoea patients, 148 (4·84%) were positive for C. jejuni. The incidence of C. jejuni in hospitals A, B and C was 5·16% (96/1859), 4·27% (46/1078) and 4·84% (6/124), respectively (Table 2), but the difference between the hospitals was not significant. Of 2191 outpatients, 138 (6·30%) were positive for C. jejuni, while only 10/870 samples (1·15%) from ward patients were positive (Table 2). It was noteworthy that the isolation rate of C. jejuni from outpatients was significantly higher than that from ward patients (P<0·01). In patients aged <7 years, 140/2601 (5·38%) were C. jejuni-positive, which was significantly higher (P<0·01) than the number in patients aged ⩾7 years (1·74%, 8/460).
Table 2. C. jejuni detected by mPCR assay in diarrhoea patients

The average positive rate of C. jejuni was 4·48% in diarrhoea patients. Outpatients and patients aged <7 years had a higher prevalence rate than that of hospitalized patients and those aged ⩾7 years, respectively (P<0·01). The detection rate of infection during June–August was higher than that of other months (P<0·05).
According to the four seasons of the year, four time periods were defined: December–February, March–May, June–August and September–November. The mean incidence of C. jejuni in these four periods was 3·08% (17/552), 1·67% (8/479), 6·29% (56/891), and 5·88% (67/1139), respectively. Statistical analysis indicated that the detection rate of C. jejuni infection during June–August was significantly higher than it was in December–February and March–May (P<0·05).
Prevalence of C. jejuni in cattle
Of the 10 farms tested, nine (90%) farms were C. jejuni-positive. Of the 1531 rectal samples collected from the 10 farms, 119 samples (7·77%) were C. jejuni-positive, with a highest positive rate of 30·91% (Table 3). Of the total of 618 rectal samples from milk cows, 31 (5·02%) were C. jejuni-positive. The highest positive rate (11·69%) was detected in farm J, and no positive results were seen in farms B, C, F and H. Out of 368 rectal samples from heifers, 32 (8·70%) were C. jejuni-positive. In these tested flocks, the highest positive rate (30·91%) was detected in flock G, and there were no positive animals in flock H.
Table 3. C. jejuni detected by mPCR assay on cattle flock farms
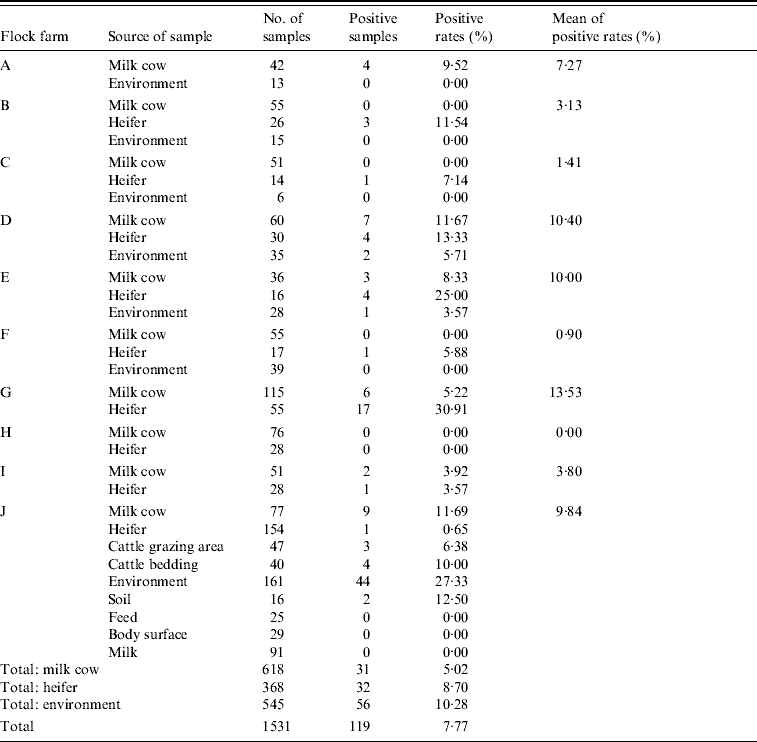
Of 1531 faecal samples collected from 10 farms, 119 (7·77%) were positive for C. jejuni, and of the 10 farms tested, six (60%) were positive for C. jejuni. The highest positive rate was up to 30·91%.
Of the 545 soil and water samples from cattle farms, 56 (10·28%) were positive for C. jejuni. In the seven farms tested, three were C. jejuni-positive (42·86%). The highest contamination rate (12·96%) was detected in farm J, and no C. jejuni was detected in farm A, B, C or F.
Prevalence of C. jejuni in poultry
Out of the 30 poultry flocks, 26 (86·67%) were C. jejuni-positive. Of 3132 chickens, 583 (18·61%) were positive for C. jejuni. The prevalence of C. jejuni varied from 0% to 73·33% in chickens from different flocks (Table 4). The positive rate for grandparent, parent and commercial chickens was 28·71% (244/850), 13·00% (52/400) and 18·03% (245/1359), respectively. Statistical analysis indicated that the C. jejuni-positive rate of grandparent chickens was significantly higher than that of parent and commercial chickens (P<0·05). In total, 541/2609 (20·74%) farm chickens were C. jejuni-positive, while only 42/523 (8·03%) of free-range chickens raised in the countryside were C. jejuni-positive. A difference in C. jejuni prevalence was also observed in different breeds, such as Ai-jiao (73·33%), Nong-da (51·30%), Luo-man (33·90%), Xin pu-dong (33·33%), and Guan-xi (37·33%) (Table 4). In addition, 6/217 (2·76%) environmental samples were C. jejuni-positive (Table 4).
Table 4. C. jejuni detected by mPCR assay on chicken flock farms
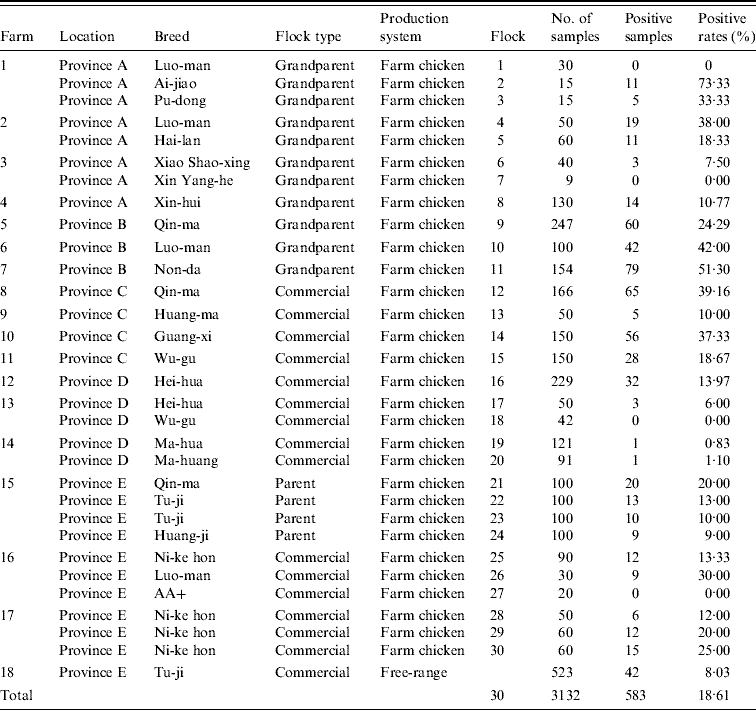
Thirty poultry flocks were investigated. Twenty-six flocks were C. jejuni-positive, with a contamination rate of 86·67%. A total of 3132 cloacal swabs were collected, 583 samples were positive for C. jejuni, and the average positive rate was 18·61%. The percentage of C. jejuni-positive samples in different flocks varied from 0% to 73·33%. In addition, 6/217 (2·76%) environmental samples were positive for C. jejuni.
fla-PCR–RFLP
fla-PCR–RFLP was reproducible with a high degree of discrimination (Fig. 1). Samples 1, 3 and 7 of the 10 C. jejuni isolates had one band type; samples 4, 5 and 8 had another band type; and samples 2, 6, 9 and 10 had one other type each.
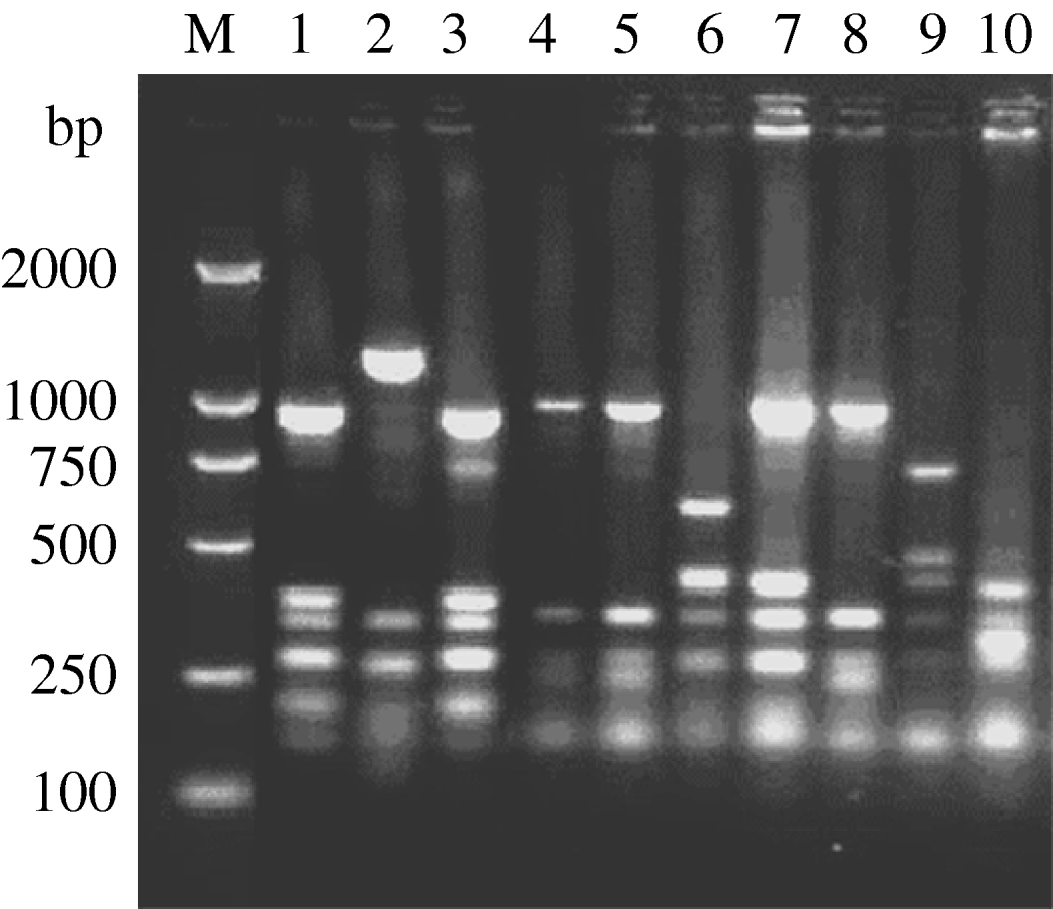
Fig. 1. Typability of fla-PCR–RFLP for C. jejuni. Lane M, DNA marker DL2000. Lanes 1–10, PCR–RFLP profiles of C. jejuni isolates from different sources digested with restriction enzyme DdeI.
A total of 315 C. jejuni isolates, including 79 from diarrhoea patients, 35 from cattle and 201 from poultry, were analysed by fla-PCR–RFLP. Two hundred and sixty-five isolates produced a flaA-PCR amplicon of the expected 1·7-kb size, which accounted for 84·13% of the total, but the others remained untypable by this method, although all of them were confirmed as C. jejuni by mPCR. The highest amplified rate (70/79, 88·61%) was observed in human isolates and the lowest rate (25/35, 71·43%) in cattle isolates (Table 5). The 265 isolates, including 70 from patients, 25 from cattle and 170 from chickens, were classified into 20 distinct fla-PCR–RFLP profiles (Fig. 2), of which profiles 18 (12·83%), 20 (10·56%) and 5 (10·19%) accounted for >10% of the total isolates each. Furthermore, profiles 2, 4, 5, 7, 8, 11–14, 16, 18 and 20 were present in all three sample sources, accounting for 72·07% of the total isolates (Table 6).
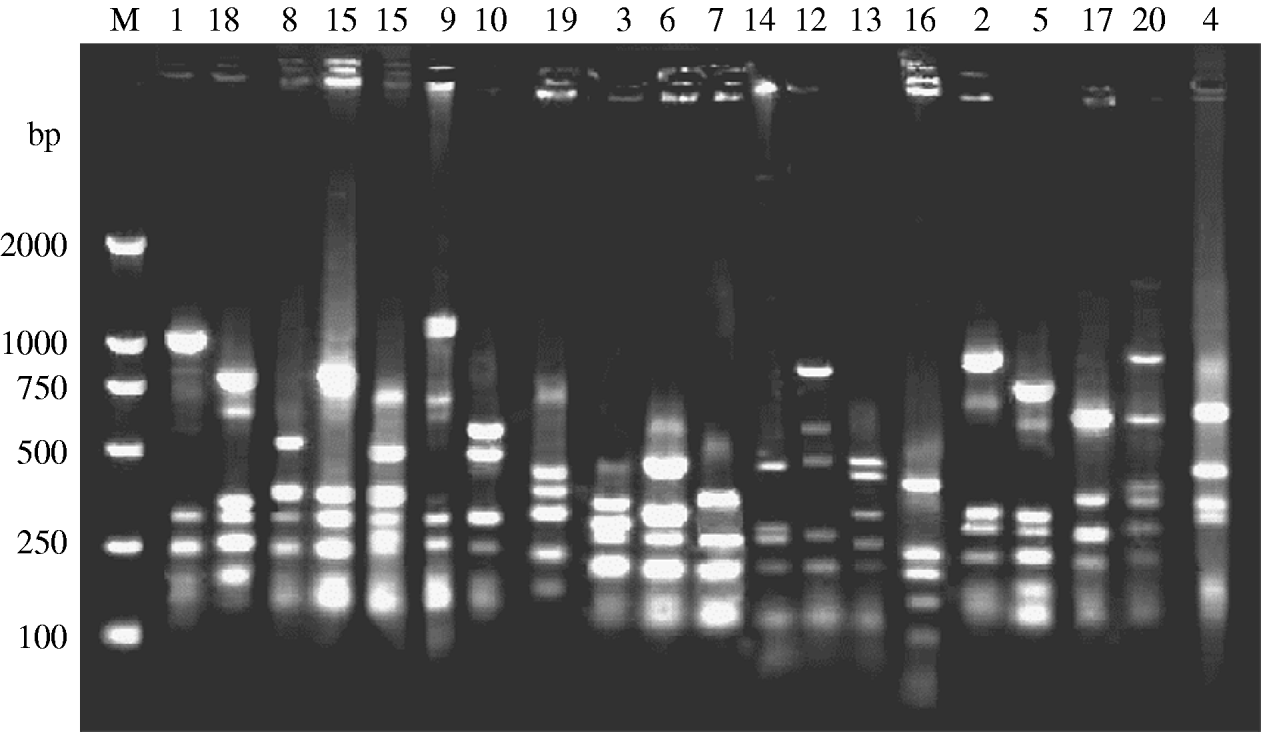
Fig. 2. fla-PCR–RFLP profiles of C. jejuni. Lane M, DNA marker DL2000. Lanes 1–20, fla-PCR–RFLP profiles of selected isolates, showing 20 kinds of profile types using restriction endonucleases DdeI.
Table 5. flaA amplification rates of C. jejuni isolates from different sources
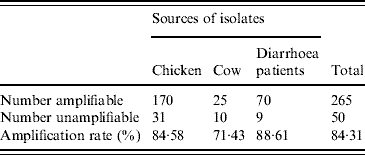
A total of 315 C. jejuni isolates, 79 from diarrhoea patients, 35 from cattle and 201 from poultry, were analysed by fla-PCR–RFLP. Two hundred and sixty-five isolates produced a flaA-PCR amplicon, which accounted for 84·13%. The amplified rate was the highest in human (88·61%) and lowest in cattle (71·43%) isolates.
Table 6. fla-PCR–RFLP analysis of C. jejuni isolates from different sources
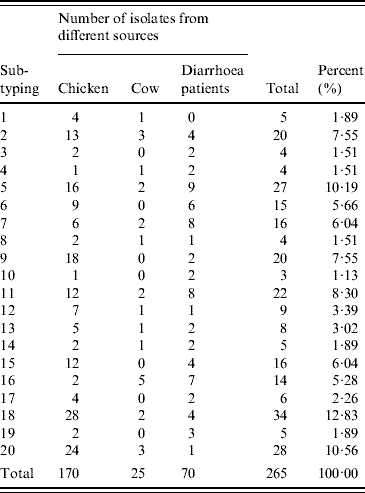
The 265 C. jejuni isolates were classified into 20 distinct profiles in which profiles 2, 5, 6, 7, 9, 11, 15, 16, 18 and 20 were the most frequently observed in different sources, and accounted for 82·67%. Of 70 human isolates, 19 different flaA types were identified and profile numbers 5, 6, 7, 11 and 16 were the most common, accounting for 54·29%. Twenty-five cattle isolates were divided into 13 different profiles. Twenty distinct profiles were identified in 170 poultry isolates. Data presented here suggested that 12 flaA types were found in human, cattle and poultry isolates, which accounted for 72·07%. The results indicated that flaA-type distribution in human isolates overlapped with that in poultry and cattle isolates. This suggests that poultry, as well as cattle, may serve as a source of human campylobacteriosis.
Out of 70 human isolates, 19 different flaA types were identified and profile 5 was the most dominant, and accounted for 10·19% of all the human isolates. No isolates of type 1 were found. Out of 25 cattle isolates, 13 different profiles were defined, and no isolates of flaA types 3, 6, 9, 10, 15, 17 or 19 were detected. Out of 170 poultry isolates, 20 distinct profiles were identified, and profiles 9, 18 and 20 were the three dominant flaA types, and accounted for 10·59, 16·47 and 14·12% of all the poultry isolates, respectively.
DISCUSSION
Infection with Campylobacter spp. is the leading cause of bacterial diarrhoea worldwide and the most common cause of travellers' diarrhoea [Reference Lang, Allos and Blaser11, Reference Tom and Frieda12]. Estimated incidence of human campylobacteriosis in industrialized countries varies from 21·9/100 000 (USA) to 396/100 000 (New Zealand) [Reference de Zoete, van Putten and Wagenaar13]. It is estimated that almost all the children in developing countries have been infected once in their lifetime with Campylobacter spp. [Reference Wheeler14]. Campylobacter can be isolated from the stool for several weeks after clinical symptoms have disappeared [Reference Wheeler14]. Therefore, children infected with C. jejuni may become sources for transmission. Perhaps the most notable complication of Campylobacter infection is GBS, an important sequela of infection with C. jejuni strains of specific serotypes [Reference Godschalk3].
The positive rate for C. jejuni in this study was similar to Oyofo et al.'s report that 4·4% of stools were positive for C. jejuni in 6760 Indonesian patients with debilitating diarrhoea [Reference Oyofo15]. From the similar positive rate found in the present study, we assumed that there is a constant positive rate of C. jejuni in humans. However, we found that outpatients had a higher positive rate than ward patients (P<0·05). This may be because most of the outpatients did not receive any antibiotics, while the ward patients were treated with appropriate antibiotics, which resulted in a lower isolation rate. However, other factors may also have affected the detection results, such as symptom severity and disease course. In a subsequent study, we will choose appropriate patient groups in order to conduct a more in-depth investigation.
The isolation rate of C. jejuni from children aged <7 years was significantly higher than that of those aged ⩾7 years. This is consistent with Oyofo et al.'s report [Reference Oyofo15], which showed that 80% of isolates (21/26) were from children aged <5 years. Our results also displayed a significant seasonal fluctuation, with a high isolation rate during June–August, and a lower isolation rate from December–February and from March–May. This was probably due to climatic differences. Numerous outbreaks of campylobacteriosis have been reported to be caused by torrential rain. For example, in Ontario, a large reported outbreak of campylobacteriosis was linked to the consumption of water from a municipal well [Reference Clark16]. Epidemiological analysis indicated that the possible source of contamination of the well was runoff from livestock following heavy rain. Bird droppings and animal waste may also be contributory factors to the contamination of surface water [Reference Whyte, McGill and Collins17].
Campylobacter is frequently isolated from poultry and cattle, therefore effective measures to reduce or eliminate carriage of C. jejuni in domestic and wild animals, especially animals raised for human consumption, are essential for intervention and control of human campylobacteriosis [Reference Hood, Pearson and Shahamat8]. About 7% of samples were positive for C. jejuni in 1531 cattle swabs in the present study, which is in agreement with the data from Dodson & LeJeune, who showed that 7% (48/686) of faecal samples from cull dairy cows was positive for C. jejuni [Reference Dodson and LeJeune6]. However, our results on the detection rate of C. jejuni in cattle were lower than those from the surveillance conducted by Ono & Yamamoto who indicated that the rate of isolation of C. jejuni from Japanese Holstein cattle was 30·9% [Reference Ono and Yamamoto18]. In Australia, there is a much wider range of carriage rates, where a median of 58% (range 12–92%) of feedlot beef cattle were C. jejuni-positive [Reference Bailey19].
Our results in poultry indicated that C. jejuni was recovered from 18·61% (583/3132) of faecal samples. Prevalence of C. jejuni in poultry flocks showed considerable variation and complex epidemiology. The carriage levels varied widely in flocks distributed in different regions. However, only 2·76% of environmental samples were positive for C. jejuni, significantly lower than that of faecal samples. The reason for the much lower prevalence of environmental samples is probably the poor adaptive response of C. jejuni to environmental stress [Reference Senok20, Reference Sonnevend21]. The differences in carriage rates in different flocks in the present study are likely to have resulted from the periodicity of the flock raised. We also observed that different raising patterns influenced greatly the carriage rates in poultry flocks. Poultry raised on a large scale makes cross-contamination much easier. In addition, the carriage rates of different breeds showed significant variation, but the reason for this is under further investigation.
A wide range of phenotypic and genotypic typing systems have been developed and used for epidemiological typing of Campylobacter [Reference Nielsen22]. To detect epidemiological linkages in isolates from different sources, a typing method with high discriminatory power is extremely important. However, routine public health surveillance of Campylobacter spp. requires a high-throughput, low-cost method with a level of discrimination that is capable of distinguishing outbreak isolates from those occurring sporadically. Of these methods, PCR–RFLP is considered especially attractive because of its sufficient discrimination, reproducibility, relative ease of use, no requirement for specific equipment, and potential application in large-scale investigations. In addition, the results can be obtained within 24 h [Reference Nachamkin, Bohachick and Patton10]. The key to this typing method lies in the primers designed for amplification and restriction enzymes selected for digestion. In the present study, 315 isolates confirmed as C. jejuni were analysed by this typing method by amplifying the flaA gene and digesting the amplified product with a single enzyme DdeI, which has been confirmed as providing the best discrimination [Reference Nachamkin, Bohachick and Patton10]. In all the isolates, 84·13% were successfully typed and classified into 20 distinct profiles. Profiles with the same band types as profiles 2, 4, 6, 7, 10, 11 and 16 in the present study were also reported by Petersen & On who indicated that PCR-Fla typing of the 30 isolates yielded 16 distinct genotypes [Reference Petersen and On23]. Nielsen et al. used six typing methods on 80 C. jejuni isolates from poultry, cattle and sporadic human clinical cases [Reference Nielsen22]. They indicated that the dominant flaA types in humans were also the most common types in cattle isolates, such as serotype 2, which represented 29% and 40% of human and cattle isolates, respectively, but only 5% of isolates from poultry. A study conducted by Oberhelman et al. has also shown that a limited number of C. jejuni that colonize chickens can cause disease in humans [Reference Oberhelman24]. Taken together, the findings of the present and previous studies indicate that certain strains of C. jejuni can circulate between humans and domestic animals such as cattle and poultry. This raises public health concerns about the risk of campylobacteriosis.
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China for Distinguished Young Scholars (30425031), Hi-tech Research and Development Program of China (2007AA02Z419), and Grants from Jiangsu Provincial Government (BK2008011, BE2008655).
DECLARATION OF INTEREST
None.












