Virus outbreaks are nothing new in history, such as the 1918 flu pandemic and the more recent Zika (2015–2016) and Ebola (2014–2016) outbreaks. But the new coronavirus-induced disease, COVID-19, is dominating the news today, since the World Health Organization (WHO) has declared COVID-19 a pandemic.
While biologists and medical researchers are working on a treatment, other scientific areas can also contribute to stopping COVID-19 and caring for patients. For example, the need for medical equipment has urged engineers to use three-dimensional (3D) printing to fabricate reanimation machines, valves, and respiratory aids, demonstrating the potential and benefits of such a technology. Materials science can play a crucial role too.
Characterizing the virus and its modes of action
Developing adequate means for treating viral infection requires prior knowledge of the microbe structure and mode of action. The entry and infection of a cell by a virus is a multistep process; the first step is the attachment of the virus to receptors at the surface of a cell. What is at the surface of the virus therefore plays a key role in the process. It is known that coronaviruses are round shells of proteins that protect the genetic material, RNA, within. On those shells are exposed spike proteins (S) that contain the receptor-binding site to the cell.Reference Seah, Su and Lingam1
Laboratory technician working with sample of coronavirus. Credit: AdobeStock.
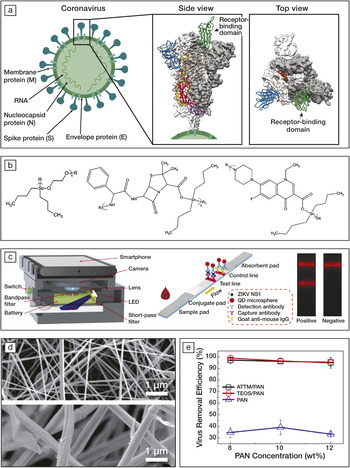
Recently, the structure of the S-proteins of COVID-19 was revealed using cryogenic electron microscopy (cryo-EM) methods (Figure 1a).Reference Wrapp, Wang, Corbett, Goldsmith, Hsieh, Abiona, Graham and McLellan2 To realize this, 3207 micrographs were taken and combined into a 3D reconstructed model with a resolution of 0.35 nm. The results show that a stochastic conformation change of the S-protein moves the receptor-binding site to an upward position, making it accessible for binding to the cell. This information could be used to develop an antiviral drug using a molecule that binds to the receptor or prevents the conformation change.
Figure 1. (a) Schematics of a coronavirus and 3D reconstructed views of the structure of its spike protein (S) showing a trimer structure. White or gray areas show the electron density obtained using cryogenic electron microscopy (cryo-EM) and colored areas represent protein conformations, where green corresponds to the receptor-binding domain. Reprinted with permission from References 1 and 2. © 2020 Springer Nature and © 2020 AAAS, respectively. (b) Repeat units of organotin polymers acting as antiviral agents. R and R1 represent side chains. Reprinted with permission from Reference 4. © 2019 Springer Nature. (c) Schematics of a smartphone-based imaging device and quantum-dot (QD)-based fluorescent reader developed for Zika virus detection, with one stripe indicating its presence (test line) and one stripe indicating the test is working (control line using an established antibody; goat anti-mouse IgG). Reprinted with permission from Reference 6. © 2019 Elsevier. (d) Electron micrographs of electrospun membranes containing 8 wt% polyacrylonitrile (PAN, top) and 10 wt% ammonium tetrathiomolybdate (ATTM/PAN, bottom) and (e) the virus removal efficiency as a function of the PAN concentration. TEOS stands for tetraethylorthosilicate. Reprinted with permission from Reference 7. © 2019 Elsevier.
After attachment and entry into a cell, viruses typically release their genetic material, use the cell machinery to replicate it, reassemble their shell around the fresh replicated DNA or RNA, and exit. Blocking any of these steps would prevent the infection, but it has been challenging to image the processes inside the cell once the virus has entered. A study showed that entire cells and their interiors could be imaged in 3D with a resolution of 8 nm.Reference Hoffman, Shtengel, Xu, Campbell, Freeman, Wang, Milkie, Pasolli, Iyer, Bogovic, Stabley, Shirinifard, Pang, Peale, Schaefer, Pomp, Chang, Lippincott-Schwartz, Kirchhausen, Solecki, Betzig and Hess3 To achieve this, a cell was first labeled with fluorescent markers, vitrified by high-pressure freezing to preserve its structure, and imaged by cryogenic light microscopy. This provided a chemical mapping of the cell. The frozen specimen was then mounted onto a cryogenic scanning electron microscope stage and milled selectively with a focused ion beam to image the topography of the interior of the cell. The images gathered from the two microscopic methods were then combined to create the 3D tomographic view of the cell. Applying this method to infected cells could reveal the mode of action of the virus inside the cell and open further avenues for drug development.
See also the MRS Bulletin December 2019 theme on Cryogenic Electron Microscopy in Materials Science
Developing antiviral drugs and effective treatments
In the current absence of a vaccine, the symptoms of COVID-19, which are similar to those of pneumonia and other forms of influenza, are being tackled. Viral infections can be treated by drugs made of molecules with special binding properties. For example, water-soluble polymers containing metals and aromatic rings can interact with the receptor-binding sites of the virus and prevent its entry inside the cell. Some of those polymers, termed organotin polymers, can also bind to the viral DNA or RNA inside the cell, preventing replication of the virus. Three examples of the repeat units of such polymers are presented in Figure 1b. The three structures derive from acyclovir, a molecule that is already included in treatments against herpes viruses. It was found that under the form of an organotin polymer, the antiviral efficiency of acyclovir could be increased and used also against other viruses, such as Zika.Reference Roner, Carraher, Miller, Mosca, Slawek, Haky and Frank4 Could it also be an effective treatment for infections by other viruses?
See also the MRS Bulletin November 2006 theme on Pharmaceutical Materials Science
Detecting the virus and diagnosing patients at an early stage
The first step to diagnose patients is to look at the symptoms and the contact history with other infected patients. However, the incubation time for a virus may reach several days during which the infected patient has no symptoms but can still spread the disease. It is crucial to develop an early stage detection method that is precise, reliable, easy, fast, and cheap. This is challenging since the most accurate and low detection method uses a polymerase chain reaction (PCR), a technique that takes one DNA or RNA segment and synthesizes a billion copies of it. This method however requires precise temperature control and expensive equipment.
To tackle this challenge and allow miniaturization for affordable point-of-care devices, many relevant materials issues can be explored. For example, in lab-on-a-chip devices conducting PCR, phase-changing materials can be used instead of batteries. Indeed, batteries are needed to power the electronic components that will control and maintain the isothermal temperature of 60°C required for the PCR. Instead, a self-heating, self-regulating passive system can be developed by exploiting the heat released during the phase change of a metallic alloy like Mg-Fe. When a drop of water is placed on the metal, an exothermic chemical reaction occurs producing temperatures in the range required for the PCR. This metal was used in a PCR chip of a few millimeters only.Reference Liao, Peng, Mauk, Awasthi, Song, Friedman, Bau and Liu5
See also the MRS Bulletin February 2006 theme on Materials for Micro- and Nanofluidics and the September 2013 theme on Quantum Dot Light-Emitting Devices
Furthermore, visual read-out is the most convenient way to see the result from point-of-care devices, similar to pregnancy tests. Fluorescent dyes are commonly used to generate this optical output. Typically, a fluorescent molecule is attached to an antibody that is specific to a biomarker of the virus. In the presence of the virus, the antibodies bind, tagging the sample with fluorophores; in absence, they are washed away. Upon shining light to the samples, the bound fluorophores emit light, indicating the presence of the virus. However, one major drawback is that a large concentration of fluorophores is required to give a strong and reliable signal, hindering the miniaturization and the detection limit.
As an alternative, nanometric quantum dots (QDs) can be used. Indeed, QDs are monodisperse nanoparticles that can emit extremely intense fluorescence. In one study, by assembling QDs into microspheres in chip-size immunoassays, Zika viruses were detected in blood samples at concentrations of 0.15 ng/mL and in just 20 minutes (Figure 1c).Reference Rong, Wang, Sun, Jia, Wang, Xiao and Wang6 Although QDs fluoresce in a narrow spectrum, they can be excited by a light with a broader spectrum. This means a smartphone could be used to induce a signal, its camera to record it, and its processing power to compute the information. Such a smartphone-based diagnosis method is cheap, portable, user-friendly, and could be a way to proceed with early diagnosis of patients.
Decontaminating and avoiding transmission
Wearing a mask (especially for healthcare professionals) and washing hands correctly and regularly are current recommendations to avoid contracting the virus. COVID-19 is transmissible when it is in the form of an aerosol, that is, liquid droplets in air. However, facemasks have limited pore size and time efficiency, and decontamination of surfaces and hands using chemicals may irritate and weaken the physical barrier made by the skin. It is therefore necessary to engineer membranes that can filter the virus and its infective form outside a host cell over a long time and in hot and humid environments such as occurs when breathing into a facemask.
In addition, to be used as a facemask, the membrane needs to be strong, flexible, enable air flow, and ideally be affordable, comfortable, and recyclable. An ideal method for fabricating defect-free and highly interconnected porous membranes on an industrial scale is electrospinning. During electrospinning, a high-voltage electric field is applied to a polymer solution to form threads that are collected onto a target. In a study using mixtures of polyacrylonitrile with tetraethoxysilane and ammonium tetrathiomolybdate salts, membranes were developed that could block more than 98% of Semliki forest virus, another RNA virus of ∼50-nm diameter (Figure 1d).Reference Al-Attabi, Rodriguez-Andres, Schütz, Bechelany, des Ligneris, Chen, Kong, Morsi and Dumee7 The efficiency of the membranes was improved by the presence of the salts that increased the surface energy and roughness of the individual polymeric fibers, enabling electrostatic binding of the virus in addition to the physical barrier provided by the mesh. Since the size of coronaviruses typically ranges from 50 nm to 200 nm in diameter, exploration of electrospun membranes may yield the next generation of facemasks.Reference Chen, Zhou, Dong, Qu, Gong, Han, Qiu, Wang, Liu, Wei, Xia, Yu, Zhang and Zhang8
See also the MRS Bulletin January 2008 theme on Advancing Materials and Technologies for Water Purification
In addition
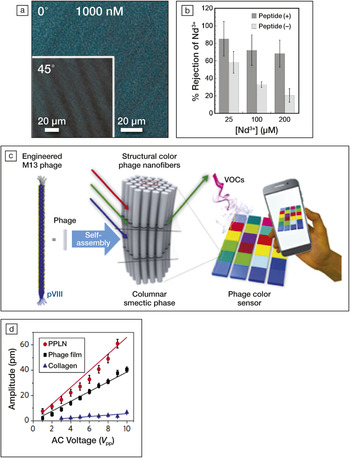
Materials science, then, can clearly contribute to fighting against COVID-19 in many ways. Inversely, virology can also contribute to the development of new materials. Indeed, viruses are not only monodisperse, with diverse and complex shapes, and with specific affinity to living organisms, they can also self-assemble and exhibit functional properties other than biological properties. For example, cylindrical M13 bacteriophages can self-assemble into flexible membranes to filter toxic ions of very small dimensions, like Nd3+ (Figure 2a–b).Reference Sawada, Inomata and Serizawa9 These self-assembled structures could be used as a template to develop new liquid crystal materials for color-based sensors (Figure 2c)Reference Lee, Fan, Samdin, Monteiro, Desai, Scheideler, Jin, Kim and Lee10 or even biodegradable piezoelectric biomaterials (Figure 2d).Reference Lee, Zhang, Zueger, Chung, Yoo, Wang, Meyer, Ramesh and Lee11 Virus-based materials engineering also offers tremendous potential for targeted cargo delivery of drugs and imaging agents, but immunogenicity remains a challenge.Reference Wu, Wu, Nakagawa and Gao12 The current urgent focus on COVID-19 could be an opportunity for further scientific and engineering breakthroughs.
Figure 2. (a) Polarized optical microscope image of self-assembled M13 bacteriophages membranes at 1000-nM concentration in virus at 0° and 45° rotation of the polarizers. Reprinted with permission from Reference 9. © 2019 Elsevier. (b) Rejection fraction of ion Nd3+ as a function of the bacteriophages concentration in the membrane for peptide-modified and unmodified viruses. Reprinted with permission from Reference 9. © 2019 Elsevier. (c) Schematics of an optical sensor using M13 self-assembly. VOCs, volatile organic chemicals. Reprinted with permission from Reference 10. © 2017 American Chemical Society. (d) Comparison of the piezoelectric response of films using M13 viruses, periodically poled lithium niobate (PPLN), and collagen. Reprinted with permission from Reference 11. © 2012 Springer Nature.
See also the MRS Bulletin February 2019 theme on Bioinspired Far-from-Equilibrium Materials




Virus outbreaks are nothing new in history, such as the 1918 flu pandemic and the more recent Zika (2015–2016) and Ebola (2014–2016) outbreaks. But the new coronavirus-induced disease, COVID-19, is dominating the news today, since the World Health Organization (WHO) has declared COVID-19 a pandemic.
While biologists and medical researchers are working on a treatment, other scientific areas can also contribute to stopping COVID-19 and caring for patients. For example, the need for medical equipment has urged engineers to use three-dimensional (3D) printing to fabricate reanimation machines, valves, and respiratory aids, demonstrating the potential and benefits of such a technology. Materials science can play a crucial role too.
Characterizing the virus and its modes of action
Developing adequate means for treating viral infection requires prior knowledge of the microbe structure and mode of action. The entry and infection of a cell by a virus is a multistep process; the first step is the attachment of the virus to receptors at the surface of a cell. What is at the surface of the virus therefore plays a key role in the process. It is known that coronaviruses are round shells of proteins that protect the genetic material, RNA, within. On those shells are exposed spike proteins (S) that contain the receptor-binding site to the cell.Reference Seah, Su and Lingam1
Laboratory technician working with sample of coronavirus. Credit: AdobeStock.
Recently, the structure of the S-proteins of COVID-19 was revealed using cryogenic electron microscopy (cryo-EM) methods (Figure 1a).Reference Wrapp, Wang, Corbett, Goldsmith, Hsieh, Abiona, Graham and McLellan2 To realize this, 3207 micrographs were taken and combined into a 3D reconstructed model with a resolution of 0.35 nm. The results show that a stochastic conformation change of the S-protein moves the receptor-binding site to an upward position, making it accessible for binding to the cell. This information could be used to develop an antiviral drug using a molecule that binds to the receptor or prevents the conformation change.
Figure 1. (a) Schematics of a coronavirus and 3D reconstructed views of the structure of its spike protein (S) showing a trimer structure. White or gray areas show the electron density obtained using cryogenic electron microscopy (cryo-EM) and colored areas represent protein conformations, where green corresponds to the receptor-binding domain. Reprinted with permission from References 1 and 2. © 2020 Springer Nature and © 2020 AAAS, respectively. (b) Repeat units of organotin polymers acting as antiviral agents. R and R1 represent side chains. Reprinted with permission from Reference 4. © 2019 Springer Nature. (c) Schematics of a smartphone-based imaging device and quantum-dot (QD)-based fluorescent reader developed for Zika virus detection, with one stripe indicating its presence (test line) and one stripe indicating the test is working (control line using an established antibody; goat anti-mouse IgG). Reprinted with permission from Reference 6. © 2019 Elsevier. (d) Electron micrographs of electrospun membranes containing 8 wt% polyacrylonitrile (PAN, top) and 10 wt% ammonium tetrathiomolybdate (ATTM/PAN, bottom) and (e) the virus removal efficiency as a function of the PAN concentration. TEOS stands for tetraethylorthosilicate. Reprinted with permission from Reference 7. © 2019 Elsevier.
After attachment and entry into a cell, viruses typically release their genetic material, use the cell machinery to replicate it, reassemble their shell around the fresh replicated DNA or RNA, and exit. Blocking any of these steps would prevent the infection, but it has been challenging to image the processes inside the cell once the virus has entered. A study showed that entire cells and their interiors could be imaged in 3D with a resolution of 8 nm.Reference Hoffman, Shtengel, Xu, Campbell, Freeman, Wang, Milkie, Pasolli, Iyer, Bogovic, Stabley, Shirinifard, Pang, Peale, Schaefer, Pomp, Chang, Lippincott-Schwartz, Kirchhausen, Solecki, Betzig and Hess3 To achieve this, a cell was first labeled with fluorescent markers, vitrified by high-pressure freezing to preserve its structure, and imaged by cryogenic light microscopy. This provided a chemical mapping of the cell. The frozen specimen was then mounted onto a cryogenic scanning electron microscope stage and milled selectively with a focused ion beam to image the topography of the interior of the cell. The images gathered from the two microscopic methods were then combined to create the 3D tomographic view of the cell. Applying this method to infected cells could reveal the mode of action of the virus inside the cell and open further avenues for drug development.
See also the MRS Bulletin December 2019 theme on Cryogenic Electron Microscopy in Materials Science
Developing antiviral drugs and effective treatments
In the current absence of a vaccine, the symptoms of COVID-19, which are similar to those of pneumonia and other forms of influenza, are being tackled. Viral infections can be treated by drugs made of molecules with special binding properties. For example, water-soluble polymers containing metals and aromatic rings can interact with the receptor-binding sites of the virus and prevent its entry inside the cell. Some of those polymers, termed organotin polymers, can also bind to the viral DNA or RNA inside the cell, preventing replication of the virus. Three examples of the repeat units of such polymers are presented in Figure 1b. The three structures derive from acyclovir, a molecule that is already included in treatments against herpes viruses. It was found that under the form of an organotin polymer, the antiviral efficiency of acyclovir could be increased and used also against other viruses, such as Zika.Reference Roner, Carraher, Miller, Mosca, Slawek, Haky and Frank4 Could it also be an effective treatment for infections by other viruses?
See also the MRS Bulletin November 2006 theme on Pharmaceutical Materials Science
Detecting the virus and diagnosing patients at an early stage
The first step to diagnose patients is to look at the symptoms and the contact history with other infected patients. However, the incubation time for a virus may reach several days during which the infected patient has no symptoms but can still spread the disease. It is crucial to develop an early stage detection method that is precise, reliable, easy, fast, and cheap. This is challenging since the most accurate and low detection method uses a polymerase chain reaction (PCR), a technique that takes one DNA or RNA segment and synthesizes a billion copies of it. This method however requires precise temperature control and expensive equipment.
To tackle this challenge and allow miniaturization for affordable point-of-care devices, many relevant materials issues can be explored. For example, in lab-on-a-chip devices conducting PCR, phase-changing materials can be used instead of batteries. Indeed, batteries are needed to power the electronic components that will control and maintain the isothermal temperature of 60°C required for the PCR. Instead, a self-heating, self-regulating passive system can be developed by exploiting the heat released during the phase change of a metallic alloy like Mg-Fe. When a drop of water is placed on the metal, an exothermic chemical reaction occurs producing temperatures in the range required for the PCR. This metal was used in a PCR chip of a few millimeters only.Reference Liao, Peng, Mauk, Awasthi, Song, Friedman, Bau and Liu5
See also the MRS Bulletin February 2006 theme on Materials for Micro- and Nanofluidics and the September 2013 theme on Quantum Dot Light-Emitting Devices
Furthermore, visual read-out is the most convenient way to see the result from point-of-care devices, similar to pregnancy tests. Fluorescent dyes are commonly used to generate this optical output. Typically, a fluorescent molecule is attached to an antibody that is specific to a biomarker of the virus. In the presence of the virus, the antibodies bind, tagging the sample with fluorophores; in absence, they are washed away. Upon shining light to the samples, the bound fluorophores emit light, indicating the presence of the virus. However, one major drawback is that a large concentration of fluorophores is required to give a strong and reliable signal, hindering the miniaturization and the detection limit.
As an alternative, nanometric quantum dots (QDs) can be used. Indeed, QDs are monodisperse nanoparticles that can emit extremely intense fluorescence. In one study, by assembling QDs into microspheres in chip-size immunoassays, Zika viruses were detected in blood samples at concentrations of 0.15 ng/mL and in just 20 minutes (Figure 1c).Reference Rong, Wang, Sun, Jia, Wang, Xiao and Wang6 Although QDs fluoresce in a narrow spectrum, they can be excited by a light with a broader spectrum. This means a smartphone could be used to induce a signal, its camera to record it, and its processing power to compute the information. Such a smartphone-based diagnosis method is cheap, portable, user-friendly, and could be a way to proceed with early diagnosis of patients.
Decontaminating and avoiding transmission
Wearing a mask (especially for healthcare professionals) and washing hands correctly and regularly are current recommendations to avoid contracting the virus. COVID-19 is transmissible when it is in the form of an aerosol, that is, liquid droplets in air. However, facemasks have limited pore size and time efficiency, and decontamination of surfaces and hands using chemicals may irritate and weaken the physical barrier made by the skin. It is therefore necessary to engineer membranes that can filter the virus and its infective form outside a host cell over a long time and in hot and humid environments such as occurs when breathing into a facemask.
In addition, to be used as a facemask, the membrane needs to be strong, flexible, enable air flow, and ideally be affordable, comfortable, and recyclable. An ideal method for fabricating defect-free and highly interconnected porous membranes on an industrial scale is electrospinning. During electrospinning, a high-voltage electric field is applied to a polymer solution to form threads that are collected onto a target. In a study using mixtures of polyacrylonitrile with tetraethoxysilane and ammonium tetrathiomolybdate salts, membranes were developed that could block more than 98% of Semliki forest virus, another RNA virus of ∼50-nm diameter (Figure 1d).Reference Al-Attabi, Rodriguez-Andres, Schütz, Bechelany, des Ligneris, Chen, Kong, Morsi and Dumee7 The efficiency of the membranes was improved by the presence of the salts that increased the surface energy and roughness of the individual polymeric fibers, enabling electrostatic binding of the virus in addition to the physical barrier provided by the mesh. Since the size of coronaviruses typically ranges from 50 nm to 200 nm in diameter, exploration of electrospun membranes may yield the next generation of facemasks.Reference Chen, Zhou, Dong, Qu, Gong, Han, Qiu, Wang, Liu, Wei, Xia, Yu, Zhang and Zhang8
See also the MRS Bulletin January 2008 theme on Advancing Materials and Technologies for Water Purification
In addition
Materials science, then, can clearly contribute to fighting against COVID-19 in many ways. Inversely, virology can also contribute to the development of new materials. Indeed, viruses are not only monodisperse, with diverse and complex shapes, and with specific affinity to living organisms, they can also self-assemble and exhibit functional properties other than biological properties. For example, cylindrical M13 bacteriophages can self-assemble into flexible membranes to filter toxic ions of very small dimensions, like Nd3+ (Figure 2a–b).Reference Sawada, Inomata and Serizawa9 These self-assembled structures could be used as a template to develop new liquid crystal materials for color-based sensors (Figure 2c)Reference Lee, Fan, Samdin, Monteiro, Desai, Scheideler, Jin, Kim and Lee10 or even biodegradable piezoelectric biomaterials (Figure 2d).Reference Lee, Zhang, Zueger, Chung, Yoo, Wang, Meyer, Ramesh and Lee11 Virus-based materials engineering also offers tremendous potential for targeted cargo delivery of drugs and imaging agents, but immunogenicity remains a challenge.Reference Wu, Wu, Nakagawa and Gao12 The current urgent focus on COVID-19 could be an opportunity for further scientific and engineering breakthroughs.
Figure 2. (a) Polarized optical microscope image of self-assembled M13 bacteriophages membranes at 1000-nM concentration in virus at 0° and 45° rotation of the polarizers. Reprinted with permission from Reference 9. © 2019 Elsevier. (b) Rejection fraction of ion Nd3+ as a function of the bacteriophages concentration in the membrane for peptide-modified and unmodified viruses. Reprinted with permission from Reference 9. © 2019 Elsevier. (c) Schematics of an optical sensor using M13 self-assembly. VOCs, volatile organic chemicals. Reprinted with permission from Reference 10. © 2017 American Chemical Society. (d) Comparison of the piezoelectric response of films using M13 viruses, periodically poled lithium niobate (PPLN), and collagen. Reprinted with permission from Reference 11. © 2012 Springer Nature.
See also the MRS Bulletin February 2019 theme on Bioinspired Far-from-Equilibrium Materials