Introduction
Proteins are among the pinnacle of polymeric materials. Nature has developed the ability to produce these monodisperse polymers with a palette of 20 amino acid monomers and nearly complete sequence control. This unparalleled level of complexity allows proteins to precisely fold and self-assemble forming not only enzymes and antibodies, but also multifunctional materials with remarkable mechanical, optical, and electronic properties. For decades, scientists and engineers have sought to understand and replicate the complex relationships between structure and processing conditions that governs the properties of these materials.Reference van Hest and Tirrell1–Reference Kan and Joshi3 This work has been carried out by studying and harvesting protein materials from native organisms and, more recently, by engineering biological systems to produce and assemble protein materials. While humans have relied on naturally harvested proteins as tools, textiles, and adhesives for thousands of years, recent advancements in biomedicine (tissue engineering, drug delivery, neural prosthetics, and wound healing) and engineered living materials, as well as the urgent need for sustainable alternatives to synthetic plastics have created an unprecedented demand for the scalable production of engineered protein-based materials with properties and functionality tailored to the specific application.
Protein engineering can be employed to encode protein-based materials with desired properties and functionality. Recent advances in synthetic biology have further accelerated the pace at which protein materials can be engineered. The design-build-test-learn loop can be used to iteratively evolve proteins with properties and functionality tailored to the desired application (Figure 1). In the design stage, artificial genes encoding modular elements, inspired by those in nature or designed de novo, can be flexibly combined to create new protein block copolymers or to outfit proteins with functional domains such as stimuli-responsive sequences, enzymes, and recognition sites. The build stage comprises of DNA assembly, cloning into organisms, protein expression, purification, and processing. Next, in the test stage, the desired materials properties are frequently measured through conventional characterization methods. This often represents a significant bottleneck as these characterization methods can be time-consuming and require a significant amount of material. Finally, the learn stage utilizes the measured data and any available computational models to provide insight into the structure–property relationship and new potential designs for future iterations.

Figure 1. Iterative design-build-test-learn loop for engineering protein materials. Recent advances in synthetic biology have enabled high throughput and iterative engineering of protein-based materials.
While the design-build-test-learn loop holds great promise, the ability to use this iterative process to engineer protein materials has lagged behind its use in other areas such as natural products, biofuels, and pharmaceuticals. This gap is caused by specific challenges that are faced in the cloning, expression, and purification of large repetitive proteins. The remainder of this article will highlight synthetic biology advancements in production hosts and cell-free systems specifically aimed at overcoming these barriers.
Production hosts and tools
Over the last decade, the steep decline in DNA sequencing cost and advent of precision genome editing have accelerated synthetic biology efforts to engineer organisms for protein and chemical production. These technological advances have hastened the exploration of genetic space and allowed engineering principles to be applied to biological systems leading to the development of a litany of genetic parts and tools designed for controlling biological processes.Reference Wang, Wei and Smolke4 Engineered parts such as synthetic promoters, induction systems, sensors, and secretion systems are being deployed in an ever-expanding number of non-model systems for the synthesis of drugs, chemicals, enzymes, and materials.Reference Sun and Alper5 Such technological catalysts are paving the way for cell factories explicitly tailored to the production of niche protein and small molecule targets allowing research to branch out from its heavy reliance on conventional expression hosts such as Escherichia coli and Saccharomyces cerevisiae. Despite clear progress, limitations remain, particularly with regard to structural proteins such as spider silk and human collagen, whose size, repetitive sequence, and post-translational processing pose challenges to recombinant production.Reference Dinjaski and Kaplan6
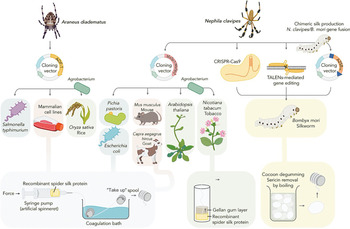
Transgenic protein expression hosts offer scalable alternatives to the native sourcing of biopolymers, such as silk and collagen, and hold immense promise for facilitating their economic synthesis from renewable feedstocks while improving batch-to-batch consistency and processing time.Reference Keating and Young7 Dragline spider silk, perhaps the holy grail of protein polymers, is one of the strongest biopolymers known and a long-sought target of recombinant production research. As native sourcing from spiders is not feasible, significant effort has been invested in porting spider silk genes into diverse hosts spanning bacterial, yeast, arthropod, mammalian, and plant systems (Figure 2).Reference Whittall, Baker, Breitling and Takano8 Of these hosts, the silkworm, Bombyx mori, is a promising vehicle for silk fiber production as it naturally spins aqueous silk protein into solid fiber. Genome editing tools such as clustered regularly interspaced palindromic repeats (CRISPR/Cas9), transposon vectors, and transcription activator-like effector nucleases (TALEN) have been utilized to deliver the spidroin gene sequence to silkworm embryos creating miniature recombinant silk extruding bioreactors.Reference Zhang, Xia, Day, Harris, Oliveira, Knittel, Licon, Gong, Dion, Lewis and Jones9–Reference Teulé, Miao, Sohn, Kim, Hull, Fraser, Lewis and Jarvis11 The silk produced in these and similar efforts is often a spider/silk protein composite exhibiting an intermediate strength and elasticity profile, although progress is advancing toward production of recombinant silk that is indistinguishable from natural dragline spider silk. Microbial silk expression systems are also promising due to their simplified culture conditions, ease of manipulation, and high yields with several companies now also attempting commercialization of these technologies.Reference Service12 The positive correlation between silk size and strength dictates that as hosts are improved for production of large recombinant proteins, the complete recapitulation of spider silk's mechanical properties will be realized.
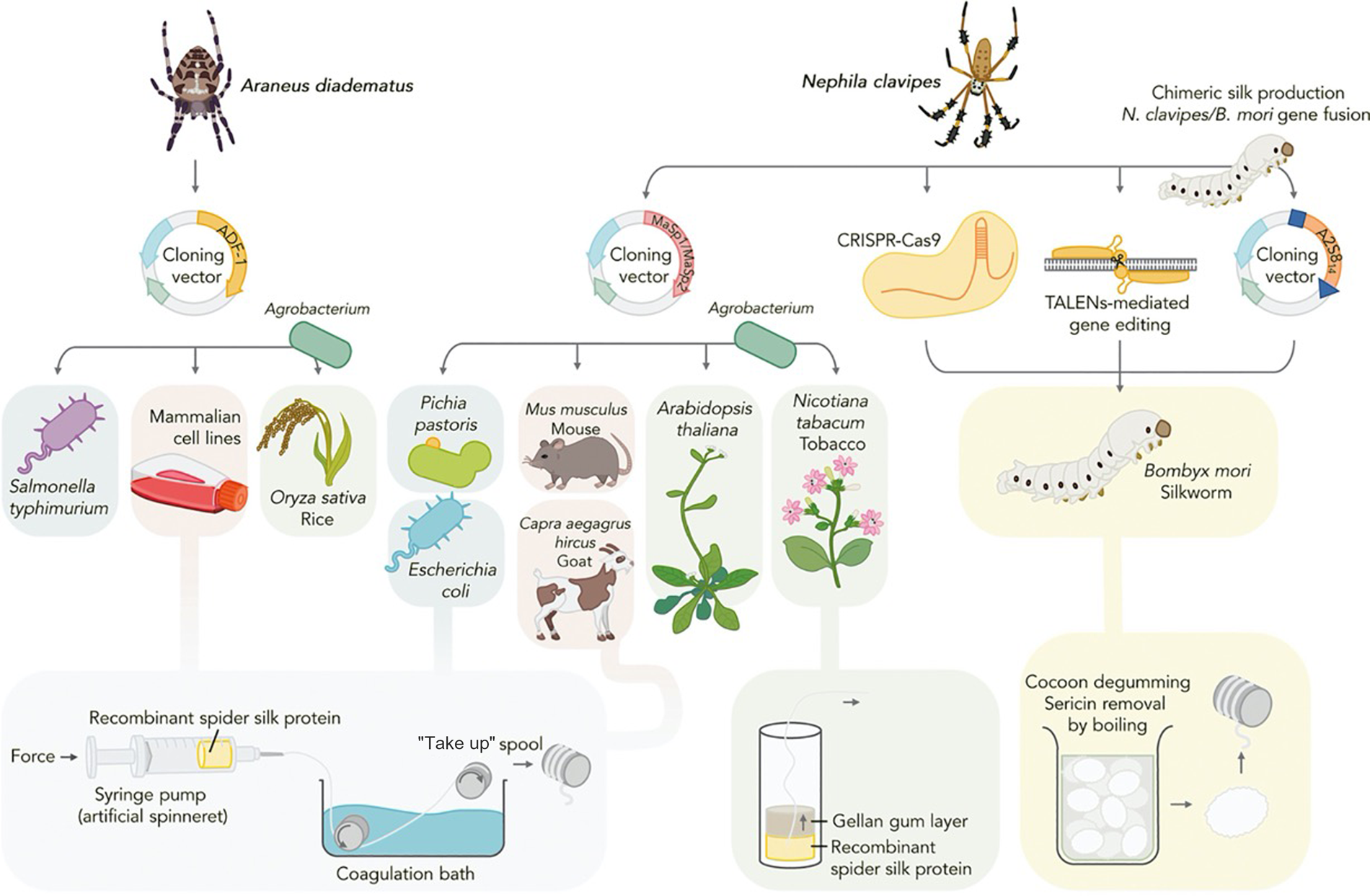
Figure 2. Expression hosts used for recombinant spider silk production. Recombinant spidroin genes derived from Nephila clavipes and Araneus diadematus have been produced in a variety of hosts spanning mammalian, insect, plant, yeast, and microbial systems. Following purification, spidroins are assembled into fibers. Transgenic silkworm, Bombyx mori, is the only production system capable of spinning cocoons containing recombinant spidroins and is amenable to a variety of gene editing techniques. Reprinted with permission from Reference 8. © 2020 Elsevier.
Principally owing to its rapid growth rate and genetic tractability, E. coli has long been the default host for protein and chemical production. Despite its prevalence, E. coli is by no means a universal platform for heterologous expression as inclusion bodies, protein toxicity, and poor expression are common with experiments suggesting roughly only one-half of bacterial proteins and less than 15% of eukaryotic proteins are stably expressed in E. coli. Reference Braun and LaBaer13 In addition to novel host development, genetic tools and metabolic engineering will help circumvent intractable expression of foreign genes in model systems. One such approach uses split inteins to assemble smaller, manageable expression units that are subsequently purified and assembled in vitro. As inteins are self-splicing peptide domains, they can be fused to bacterially expressed proteins of interest for in vitro assembly. Silk domains have been expressed with intein tags enabling ligation of spidroin silk domains yielding >500 KDa oligomers that retain the mechanical properties of natural dragline silk.Reference Bowen, Dai, Sargent, Bai, Ladiwala, Feng, Huang, Kaplan, Galazka and Zhang14 Split inteins have similarly been used for the assembly of trimers of the adhesive mussel foot protein mfp5 to overcome poor protein yields.Reference Kim, Dai, Qiao, Li, Fortner and Zhang15 Metabolic engineering of the E. coli translation apparatus and associated amino acid pool has also been shown to enhance overexpression of tRNA synthetases, cognate tRNAs, and amino acid biosynthetic enzymes aiding expression of spider silk and mussel foot proteins.Reference Jeong, Yang, Yang, Shin, Seong, Cha and Kwon16–Reference Xia, Qian, Ki, Park, Kaplan and Lee18 Such approaches serve as a roadmap for the gradual improvement of biopolymer production by employing synthetic biology tools to tailor bioproduction to the exquisite demands of specific biopolymer of interest.
Other microbial hosts such as the yeast Pichia pastoris and the bacterium Salmonella enterica possess characteristics useful for protein polymer production. Notably, P. pastoris grows to high density, encodes a versatile protein secretion system, and boasts a rapidly expanding set of genetic tools for tuning protein production.Reference Obst, Lu and Sieber19 Its secretion system is particularly useful as it can act as a first-pass purification step, is tolerant of large proteins, complements posttranslational processing, and has been used to efficiently biosynthesize and secrete silk-like polymers at g/L quantities.Reference Werten, Moers, Vong, Zuilhof, van Hest and de Wolf20 Likewise, the secretion system endogenous to S. enterica holds promise for expression of proteins such as silk, elastin, and collagen whose large repetitive sequences make them prone to aggregation and degradation.Reference Azam, Li, Metcalf and Tullman-Ercek21
Protein biopolymer production in plant systems has also borne fruit with transgenic arabidopsis, maize, tobacco, and potato expression systems all showing utility in biopolymer production. For example, the type I human collagen genes and accompanying proline and lysine hydroxylases were coexpressed in tobacco to generate a high-producing strain (~2% collagen of total protein) for human pro-collagen that exhibited a posttranslational profile highly similar to that of native collagen.Reference Stein, Wilensky, Tsafrir, Rosenthal, Amir, Avraham, Ofir, Dgany, Yayon and Shoseyov22 Critically, the recombinant collagen retained the native trihelical structure essential for self-assembling the higher-order fibrillar structure typical of extracellular collagen and also displayed increased hydrophilicity compared to native collagen enabling high concentrations potentially useful for shear-force-based fibril alignment to more closely mimic the environment and mechanics of the extracellular matrix.Reference Yaari, Posen and Shoseyov23
An important benefit of recombinant production is the relative ease with which protein sequence can be altered, with chimeric fusion proteins and site-specific variants being two common techniques to expand functionality and alter materials properties.Reference Dinjaski and Kaplan6 Such approaches are particularly useful in the design of highly mutable proteins such as elastin-like polypeptides where repeat motifs contain customizable residues that modulate protein properties such as phase-transition temperature allowing the creation of thermally responsive biomaterials. Development of chimeric recombinant structural proteins such as silk-elastin-like proteins is currently an active area of research where chimeric proteins display improved properties involved in catalysis, gelation, solubility, mineralization, and adhesion.Reference Dimarco and Heilshorn24
Beyond the cell
The tug-of-war between the objectives of bioengineers to produce and release a single biomolecular product and cell survival manifests itself in a variety of common challenges constraining the current state of the art (e.g., low product yields, product toxicity, and a limited chemical palette of potential products). Cell-free systems are emerging as a new opportunity to enable expanded biological capabilities.Reference Silverman, Karim and Jewett25 The foundational principle is that precise, complex biomolecular transformations can be conducted in crude cell lysates without intact cells. This concept circumvents mechanisms that have evolved to facilitate species survival, bypasses limitations on molecular transport across the cell wall, and provides a significant departure from traditional, cell-based processes that rely on microscopic cellular “reactors.” Two key areas in engineered protein materials that have recently emerged include: cell-free protein synthesis of biopolymers containing noncanonical amino acids and cell-free ribosome engineering. Next, we describe each of these in turn.
The extraordinary synthetic capability of nature's protein biosynthesis system, which includes the ribosome and the associated factors needed for polymerization, has driven extensive efforts to harness it for societal needs (e.g., insulin production). In nature, however, only limited sets of ribosomal monomers are utilized, thereby resulting in limited sets of biopolymers (i.e., proteins). Expanding nature's repertoire of ribosomal monomers and polymerization chemistries could yield new classes of enzymes, therapeutics, and materials with diverse genetically encoded chemistry.Reference Arranz-Gibert, Vanderschuren and Isaacs26–Reference Hammerling, Kruger and Jewett28 Recent efforts to expand the genetic code have repurposed the natural translation system to selectively incorporate more than 150 noncanonical amino acids (ncAAs) into proteins to enable a wave of exciting new applications in molecular imaging,Reference Cellitti, Jones, Lagpacan, Hao, Zhang, Hu, Brittain, Brinker, Caldwell, Bursulaya, Spraggon, Brock, Ryu, Uno, Schultz and Geierstanger29 site-specific incorporation of posttranslational modifications and their mimics,Reference Neumann, Peak-Chew and Chin30–Reference Venkat, Sturges, Stahman, Gregory, Gan and Fan32 fluorescent probes,Reference Vázquez, Nitz, Stehn, Yaffe and Imperiali33–Reference Cho, Daniel, Buechler, Litzinger, Maio, Putnam, Kraynov, Sim, Bussell, Javahishvili, Kaphle, Viramontes, Ong, Chu, Gc, Lieu, Knudsen, Castiglioni, Norman, Axelrod, Hoffman, Schultz, DiMarchi and Kimmel35 medicines,Reference Axup, Bajjuri, Ritland, Hutchins, Kim, Kazane, Halder, Forsyth, Santidrian, Stafin, Lu, Tran, Seller, Biroc, Szydlik, Pinkstaff, Tian, Sinha, Felding-Habermann, Smider and Schultz36–Reference Katoh, Sengoku, Hirata, Ogata and Suga39 and genetically encoded materials.Reference Albayrak and Swartz40–Reference Baumann, Nickling, Bartholomae, Buivydas, Kuipers and Budisa44 Underpinning such advances, platform technologies have emerged to facilitate high-level expression of proteins containing ncAAs. These technologies include the development of genomically recoded organisms where all occurrences of the amber stop codon have been genomically recoded to the ochre TAA stop codon, which permits deletion of release factor 1 (RF1) and complete reassignment of the amber codon translation function for a defined ncAA.Reference Lajoie, Rovner, Goodman, Aerni, Haimovich, Kuznetsov, Mercer, Wang, Carr, Mosberg, Rohland, Schultz, Jacobson, Rinehart, Church and Isaacs45 This is significant because it alleviates a long-standing issue of RF1 competition, which has historically led to poor protein expression yields and inefficient incorporation of multiple identical ncAAs by amber suppression. In cell-free systems, expression platforms based on these strains have enabled protein synthesis yields of up to 2.7 g/L and site-specific incorporation of up to 40 ncAAs into elastin-like polypeptides with high accuracy (≥ 98%).Reference Martin, Des Soye, Kwon, Kay, Davis, Thomas, Majewska, Chen, Marcum, Weiss, Stoddart, Amiram, Ranji Charna, Patel, Isaacs, Kelleher, Hong and Jewett46,Reference Des Soye, Gerbasi, Thomas, Kelleher and Jewett47 These are the purest polymers with this many site-specifically introduced ncAAs synthesized to date. It sets the stage for new classes of functional materials, where the basic biopolymer structure is elaborated with pendant moieties to program physical properties with atomic-scale resolution.
While site-specific incorporation of such diverse chemistries into peptides and proteins has facilitated exciting applications, numerous classes of noncanonical monomers (e.g., backbone-extended β- and γ- amino acids) remain poorly compatible with the natural translation apparatus. While exciting new innovations are occurring,Reference Katoh, Sengoku, Hirata, Ogata and Suga39,Reference Lee, Schwieter, Watkins, Kim, Yu, Schwarz, Lim, Coronado, Byrom, Anslyn, Ellington, Moore and Jewett48–Reference Lee, Schwieter, Watkins, Kim, Yu, Schwarz, Lim, Coronado, Byrom, Anslyn, Ellington, Moore and Jewett54 especially challenging constraint to incorporate nonalpha-NCAAs is the ribosome, which has evolved to polymerize α−amino acids.Reference Hammerling, Fritz, Yoesep, Kim, Carlson and Jewett55 Because the ribosome's function is necessary for life, cell viability restricts the ribosomal mutations that can be made. To address this challenge, several new efforts are emerging to build and evolve ribosomes in vitro that are decoupled from cellular growth, providing transformative opportunities to expand the chemistry of life.Reference Hammerling, Fritz, Yoesep, Kim, Carlson and Jewett55
In this issue
This issue of MRS Bulletin provides articles written by leading researchers in protein materials science. These contributions provide insights into cutting-edge developments that both further our understanding of protein materials in native systems and boost our ability to harness protein engineering for materials development. The article by Shi et al. discusses how proteins are produced through extraction from native organisms and recombinant expression in engineered hosts.Reference Shi, Pathiranage and Marciel56 The article then delves into the fundamentals of how protein amino acid sequence impacts the rheological and mechanical properties of natural and engineered proteins. Recently, proteins have been found to play a crucial role in the liquid–liquid phase separation that causes the assembly of subcellular structures such as the nucleolus. Sun et al. provide a review of recent developments in understanding how liquid–liquid phase separation is used by organisms to assemble protein-based materials and how this understanding can be leveraged in engineered systems.Reference Sun, Lim, Qi, Yu and Miserez57
In native organisms, many structural proteins are found in nanocomposites alongside inorganic materials (e.g., bone and nacre). In such systems, the protein not only serves as the matrix material, but also serves to guide the assembly or formation of the inorganic reinforcement. Wang et al. discuss advancements in the assembly of proteins with nanomaterials to create high-performance nanocomposites.Reference Wang, Kang, Cao, Krecker, Tsukruk and Singamaneni58 Moving beyond structural materials, many organisms assemble protein materials with remarkable optical and electronic properties (e.g., keratin in iridescent bird feathers and electrically conducting protein filaments in bacterial biofilms). Dennis et al. cover how natural and bioinspired proteins can be used to create electronic and optical materials.Reference Dennis, Onderko, Slocik, Bird, Phillips, Crookes-Goodson and Glaven59 The review by Iranmanesh et al. covers recent advances in protein engineering for functional nanomaterials.Reference Iranmanesh, Subhash, Glover and Bedford60
The convergence of protein engineering and materials processing methods such as additive manufacturing has led to a flood of advancements in the engineering of tissues and organs. Recently, these techniques have also been used to produce engineered living materials, where microbes are embedded in a biopolymer matrix to create materials with sense and respond functionality. The article by Gona and Meyer review the exciting developments in three-dimensional printing of engineered proteins for living materials.Reference Gona and Meyer61
Conclusion
Recombinant production of protein polymers has lagged behind that of pharmaceutical and commodity chemicals due in part to the heavy metabolic burden inherent to over-expression of large nonnative genes. Widespread success in this area depends upon overcoming multiple nontrivial metabolic obstacles in order to deliver yields high enough to compete with natively sourced material. Despite the challenge, current results demonstrate the investment is well worth the cost as these biofactories are fulfilling the promise to improve the economics and versatility of structural protein production. Future achievements are sure to impact myriad sectors ranging from tissue engineering, fabrics, and cosmetics as the ongoing effort to engineer host organisms continues to yield tangible results moving us closer to the controlled production of tailored protein polymers at the industrial scale.
 Maneesh Gupta is a materials scientist at the Air Force Research Laboratory, Materials and Manufacturing Directorate. He received his PhD degree in materials science and engineering from the Georgia Institute of Technology. He received his BS degree in chemical engineering from the University of Illinois at Urbana-Champaign. Gupta was a postdoctoral fellow at Princeton University, where he developed additive manufacturing methods for the incorporation of controlled-release capsules in three-dimensional printed tissue engineering matrices. His current research focuses on the processing and characterization of bioinspired protein-based materials, with a specific emphasis on the ability to produce materials with tunable and graded properties. Gupta can be reached by email at maneesh.gupta.2@us.af.mil.
Maneesh Gupta is a materials scientist at the Air Force Research Laboratory, Materials and Manufacturing Directorate. He received his PhD degree in materials science and engineering from the Georgia Institute of Technology. He received his BS degree in chemical engineering from the University of Illinois at Urbana-Champaign. Gupta was a postdoctoral fellow at Princeton University, where he developed additive manufacturing methods for the incorporation of controlled-release capsules in three-dimensional printed tissue engineering matrices. His current research focuses on the processing and characterization of bioinspired protein-based materials, with a specific emphasis on the ability to produce materials with tunable and graded properties. Gupta can be reached by email at maneesh.gupta.2@us.af.mil.
 Drew Wagner is a research scientist at the Air Force Research Laboratory. He obtained his BS degree in biochemistry and molecular biology from the University of California, Santa Cruz, in 2010. Upon entering graduate school at The University of Texas at Austin, he investigated the structure and function of enzymatic assembly lines involved in antibiotic biosynthesis. His research uses synthetic biology tools to study self-assembling protein complexes. Wagner can be reached by email at drew@debutbiotech.com.
Drew Wagner is a research scientist at the Air Force Research Laboratory. He obtained his BS degree in biochemistry and molecular biology from the University of California, Santa Cruz, in 2010. Upon entering graduate school at The University of Texas at Austin, he investigated the structure and function of enzymatic assembly lines involved in antibiotic biosynthesis. His research uses synthetic biology tools to study self-assembling protein complexes. Wagner can be reached by email at drew@debutbiotech.com.
 Michael C. Jewett is the Charles Deering McCormick Professor of Teaching Excellence, Walter P. Murphy Professor of Chemical and Biological Engineering, and Director of the Center for Synthetic Biology at Northwestern University. He is also an Institute Fellow at the Northwestern Argonne Institute for Science and Engineering. His research seeks to re-conceptualize the way we engineer complex biological systems for compelling applications in medicine, materials, and energy by transforming biochemical engineering with synthetic biology. His awards include the National Institutes of Health Pathway to Independence Award in 2009, the David and Lucile Packard Fellowship in Science and Engineering in 2011, and the Defense Advanced Research Projects Agency Young Faculty Award in 2011, among others. He is an associate editor for ACS Synthetic Biology. Jewett can be reached by email at m-jewett@northwestern.edu.
Michael C. Jewett is the Charles Deering McCormick Professor of Teaching Excellence, Walter P. Murphy Professor of Chemical and Biological Engineering, and Director of the Center for Synthetic Biology at Northwestern University. He is also an Institute Fellow at the Northwestern Argonne Institute for Science and Engineering. His research seeks to re-conceptualize the way we engineer complex biological systems for compelling applications in medicine, materials, and energy by transforming biochemical engineering with synthetic biology. His awards include the National Institutes of Health Pathway to Independence Award in 2009, the David and Lucile Packard Fellowship in Science and Engineering in 2011, and the Defense Advanced Research Projects Agency Young Faculty Award in 2011, among others. He is an associate editor for ACS Synthetic Biology. Jewett can be reached by email at m-jewett@northwestern.edu.