Introduction
Understanding the genetic basis and mode of inheritance of herbicide resistance is essential for predicting its persistence, evolutionary trajectory, and rate of spread in a weed species (Jasieniuk et al. Reference Jasieniuk, Brule-Babel and Morrison1996; Neve Reference Neve2007). Such information is especially valuable for resistance to glyphosate, the most widely used herbicide globally (Duke Reference Duke2018). Incidences of glyphosate resistance have increased steadily since the first case was reported in Australia in 1995 (Pratley et al. Reference Pratley, Urwin, Stanton, Baines, Broster, Cullis, Schafer, Bohn and Krueger1999), and it has now been confirmed in 41 weed species worldwide (Heap Reference Heap2018). Some glyphosate-resistance mechanisms are inherited as a dominant or semidominant allele at a single locus (Christoffers and Varanasi Reference Christoffers and Varanasi2010). Examples include glyphosate resistance based on vacuolar sequestration in horseweed (Erigeron canadensis L.) (Ge et al. Reference Ge, d’Avignon, Ackerman and Sammons2010; Zelaya et al. Reference Zelaya, Owen and Vangessel2004); resistance resulting from reduced herbicide translocation in rigid ryegrass (Lolium rigidum Gaudin) (Wakelin and Preston Reference Wakelin and Preston2006; Preston and Wakelin Reference Preston and Wakelin2008); and resistance associated with amino acid substitutions at the Pro-106 position within the EPSPS gene in several weed species, including goosegrass [Eleusine indica (L.) Gaertn.] (Ng et al. Reference Ng, Ratnam, Surif and Ismail2004) and Italian ryegrass [Lolium perenne (L). ssp. multiflorum Lam. Husnot] (Jasieniuk et al. Reference Jasieniuk, Ahmad, Sherwood, Firestone, Perez-Jones, Lanini, Mallory-Smith and Stednick2008). Simarmata et al. (Reference Simarmata, Bughrara and Penner2005) reported glyphosate resistance in a Californian L. rigidum population inherited as two semidominant independently segregating alleles. However, Yu et al. (Reference Yu, Cairns and Powles2007) described a South African population of L. rigidum with resistance to multiple herbicides, including glyphosate; these plants had accumulated independently inherited glyphosate-resistance alleles at different loci, including combining the Pro-106 EPSPS target-site mutation with reduced glyphosate translocation as an additional resistance mechanism. It is possible, therefore, that glyphosate resistance apparently controlled by a two-locus system may in fact consist of two different mechanisms, each controlled at a single locus and acting additively.
An exception to resistance mechanisms mediated at one or two loci is glyphosate resistance based on amplification of the EPSPS gene. This increase in gene-copy number results in overexpression of the EPSPS enzyme targeted by glyphosate, enabling resistant plants to produce sufficient enzyme to maintain the shikimate pathway even in the presence of glyphosate (Gaines et al. Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010; Powles Reference Powles2010). Glyphosate resistance linked to EPSPS gene amplification, first described in cell suspension cultures of tobacco (Nicotiana tabacum L.), alfalfa (Medicago sativa L.), and soybean [Glycine max (L.) Merr.] (Widholm et al. Reference Widholm, Chinnala, Ryu, Song, Eggett and Brotherton2001), was confirmed in whole plants in a Palmer amaranth (Amaranthus palmeri S. Watson) population from Georgia (Gaines et al. Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010). Since then, additional examples of this glyphosate-resistance mechanism have been reported in A. palmeri populations from North Carolina (Chandi et al. Reference Chandi, Milla-Lewis, Giacomini, Westra, Preston, Jordan, York, Burton and Whitaker2012), Mississippi (Ribeiro et al. Reference Ribeiro, Pan, Duke, Nandula, Baldwin, Shaw and Dayan2014), New Mexico (Mohseni-Moghadam et al. Reference Mohseni-Moghadam, Schroeder and Ashigh2013), and Nebraska (Chahal et al. Reference Chahal, Varanasi, Jugulam and Jhala2017). EPSPS gene amplification has also been reported in glyphosate-resistant (GR) populations of waterhemp [Amaranthus tuberculatus (Moq.) J. D. Sauer] (Chatham et al. Reference Chatham, Wu, Riggins, Hager, Young, Roskamp and Tranel2015), kochia [Bassia scoparia (L.) A. J. Scott] (Wiersma et al. Reference Wiersma, Gaines, Preston, Hamilton, Giacomini, Buell, Leach and Westra2015), L. perenne (Salas et al. Reference Salas, Dayan, Pan, Watson, Dickson, Scott and Burgos2012), ripgut brome (Bromus diandrus Roth) (Malone et al. Reference Malone, Morran, Shirley, Boutsalis and Preston2015), and windmillgrass (Chloris truncata R. Br.) (Ngo et al. Reference Ngo, Malone, Boutsalius, Gill and Preston2018). EPSPS amplification combined with the Pro-106 target-site mutation was recently reported in GR E. indica in Mexico (Gherekhloo et al. Reference Gherekhloo, Fernandez-Moreno, Alcantara de la Cruz, Sanchez-Gonzalez, Cruz- Hipolito, Dominguez-Valenzuela and De Prado2017).
Despite increasing evidence for independent evolution of GR weed populations with increased EPSPS copy numbers, few studies have examined generational transmission of EPSPS gene amplification via sexual or asexual reproduction in any weed species. Jugulam et al. (Reference Jugulam, Niehues, Godar, Koo, Danilova, Friebe, Seghal, Varanasi, Wiersma, Westra, Stahlman and Gill2014) reported that in B. scoparia, amplified EPSPS copies were present as a tandem repeat block and inherited as a single locus with Mendelian segregation. This was not the case, however, in B. diandrus, in which Malone et al. (Reference Malone, Morran, Shirley, Boutsalis and Preston2015) found variable EPSPS copy-number amplification but no segregation for glyphosate resistance in an F2 derived from a cross between resistant (R) and susceptible (S) individuals. Investigations of the inheritance of amplified EPSPS gene copies in A. palmeri have shown inconclusive results. Mohseni-Moghadam et al. (Reference Mohseni-Moghadam, Schroeder and Ashigh2013) did not find Mendelian segregation ratios consistent with a single-gene mechanism among R and S phenotypes in F1 and pseudo-F2 families. Chandi et al. (Reference Chandi, Milla-Lewis, Giacomini, Westra, Preston, Jordan, York, Burton and Whitaker2012) reported apparent single-gene segregation between R and S phenotypes in some BC1F1 families derived from crosses between R and S parents; however, these authors also found that segregation in other BC1F1 families did not conform to a single-gene model, and they suggested that results for some families also potentially conformed to a two-gene additive model. Both Mohseni-Moghadam et al. (Reference Mohseni-Moghadam, Schroeder and Ashigh2013) and Chandi et al. (Reference Chandi, Milla-Lewis, Giacomini, Westra, Preston, Jordan, York, Burton and Whitaker2012) reported no differences in progeny from reciprocal R × S and S × R crosses, and concluded this form of glyphosate resistance is under nuclear control with no maternal effects. In contrast, Ribeiro et al. (Reference Ribeiro, Pan, Duke, Nandula, Baldwin, Shaw and Dayan2014) described apparent maternal influence on progeny EPSPS gene-copy number, possibly resulting from apomixis; however, these authors were not able to confirm apomictic progeny using DNA markers. Neither Mohseni-Moghadam et al. (Reference Mohseni-Moghadam, Schroeder and Ashigh2013) nor Chandi et al. (Reference Chandi, Milla-Lewis, Giacomini, Westra, Preston, Jordan, York, Burton and Whitaker2012) quantified EPSPS gene-copy number in the F1 or pseudo-F2 progeny: the authors estimated R:S segregation ratios by exposing progeny families to screening doses of glyphosate and recording the proportion of survivors. A major limitation of this approach is that the genomes of A. palmeri plants capable of surviving field rates of glyphosate may contain from 10 to more than 100 EPSPS gene copies (Gaines et al. Reference Gaines, Shaner, Ward, Leach, Preston and Westra2011); assigning these diverse genotypes to a single R phenotype fails to detect this potentially important variation in progeny gene-copy number. Gaines et al. (Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010; Reference Gaines, Shaner, Ward, Leach, Preston and Westra2011) reported EPSPS copy numbers ranging from 1 to more than 100 in a pseudo-F2 family of 54 plants; the gene-copy number of one of these F2 plants was greater than the combined copy numbers of the F1 parents, indicating amplified EPSPS gene transmission via sexual reproduction is not merely additive, but more complex and potentially unstable. The objective of the research reported here, therefore, was to further investigate and characterize the inheritance and intergenerational stability of amplified EPSPS gene copies in A. palmeri for both sexual and asexual reproduction.
Materials and Methods
Plant Material and EPSPS Copy-Number Estimation
Parent plants for this study were grown from seed collected in 2009 from a population in Georgia, that was not controlled by field applications of glyphosate (S Culpepper, personal communication). Seeds were plated onto sterile 1% agar in 100 mm by 15 mm petri plates (25 seeds plate−1) and placed in a germination chamber set to 35 C light/30 C dark with a 12-h photoperiod. Germinated seeds with at least 1 cm of visible shoot were transferred into potting soil (Fafard® #2 SV, Conrad Fafard) in 9-cm3 plastic pots in a growth chamber set at 30 C, 75% humidity, and a 12-h photoperiod. Five grams of slow-release fertilizer (Osmocote® Smart-Release Plant Food, 19-6-12, Scotts Miracle-Gro) was added to the soil in each pot at planting. Plants were transplanted at the 2- to 4-leaf stage into 1-L pots and transferred to a Colorado State University greenhouse where they were grown under 400-W sodium halide lamps to provide a 14-h photoperiod. Temperatures were maintained at 24 C day/18 C night. All pots were spaced at least 35 cm apart on a greenhouse bench and rerandomized weekly.
All plants used as parents in controlled crosses were initially confirmed as GR or glyphosate-susceptible (GS) using the leaf-disk shikimate assay described by Shaner et al. (Reference Shaner, Nadler-Hassar, Henry and Kroger2005). Young leaf tissue was collected from individual parent plants, and total genomic DNA was extracted using DNeasy Plant Mini Kits (Qiagen) for estimation of EPSPS copy number via qPCR as described by Giacomini et al. (Reference Giacomini, Westra and Ward2014). The acetolactate synthase (ALS) gene was used as a reference, because it occurs at a known single locus within the A. palmeri genome (Gaines et al. Reference Gaines, Zhang, Wang, Bukun, Chisholm, Shaner, Nissen, Patzoldt, Tranel, Culpepper, Grey, Webster, Vencill, Sammons, Jiang, Preston, Leach and Westra2010). Relative to this ALS standard, all plants in this study classified as S based on the shikimate assay had one EPSPS gene copy per haploid genome, and all plants classified as R had 10 or more EPSPS gene copies. To test the repeatability of qPCR-based estimates of amplified EPSPS gene-copy numbers, separate qPCR runs were repeated three times on a subset of DNA samples from 23 randomly selected F1 individuals that each had at least 10 EPSPS gene copies.
Generation and Testing of F1 and Pseudo-F2 Families
Controlled crosses were carried out by enclosing one male and one female plant together in a microperforated pollination bag as soon as the first flowers appeared and before anthesis or stigma exsertion. Eleven F1 families were produced, each from a controlled cross between a separate pair of parents. Details of the parent pair EPSPS copy numbers are given in Table 1. F1 plants were germinated and grown, and EPSPS copy numbers were analyzed as previously described for 6 to 18 progeny from each F1 family. Shikimate detection using the Shaner et al. (Reference Shaner, Nadler-Hassar, Henry and Kroger2005) in vitro leaf-disk shikimate assay was also performed on all parental and F1 plants to test for association of EPSPS gene amplification with glyphosate resistance.
Table 1 EPSPS gene-copy numbers of parent plants, F1 and F2 families.
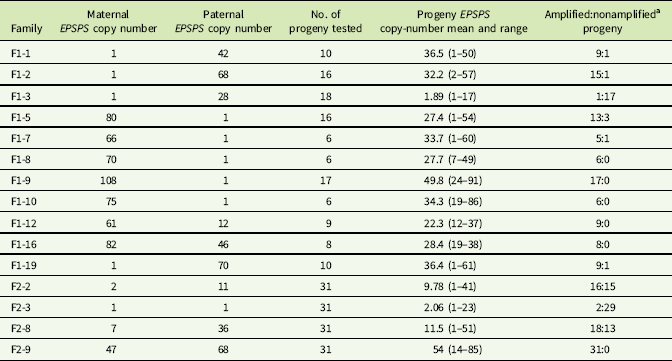
a Progeny with ≤2 estimated EPSPS copies classified as nonamplified; progeny with >2 estimated EPSPS copies classified as amplified.
Crosses between F1 plants were made to generate pseudo-F2 families by enclosing one female plant and one male plant from the same F1 family together in a microperforated pollination bag. One full-sib pair was crossed within each F1 family, and four of these pseudo-F2 families (hereafter referred to as F2 families) were selected for EPSPS copy-number examination via qPCR. Table 1 shows maternal and paternal EPSPS copy numbers for the four F2 families examined: the families were selected to represent different combinations of EPSPS copy numbers in the F1 parents (i.e., low × high, low × low, high × high). Thirty-one plants from each selected F2 family were grown under the greenhouse conditions described earlier until they were large enough for sampling of young leaf tissue for DNA extraction.
Generation and Testing of Clonally Propagated Shoots
To test the stability of EPSPS copy number in the same plant tissue over time, vegetative propagation was performed for multiple generations. Starting with a GR female plant (EPSPS copies = 122) from the original Georgia population, clones were produced by excising side shoots that were at least 15-cm long, stripping each shoot of all lower leaves to reduce transpiration loss so that 3 to 4 leaves remained at the distal end, and dipping the basal ends of the shoots in rooting hormone (RootBoost™, 0.1% indole-3 butyric acid, GardenTech). Shoots were then planted in potting soil (Fafard #2 SV, Conrad Fafard), placed under a humidity dome, and allowed to develop roots in the greenhouse under conditions of 24 C day/18 C night temperatures and a 14-h photoperiod.
After 10 successive generations of cloning using this technique, three plants were randomly selected from each of four different cloned lines, all derived from the original female plant, and qPCR EPSPS copy-number estimates were obtained for these 12 plants as previously described. A further 16 cloned individuals were then created via rooted shoot cuttings from 6 of these 12 plants, and these were also assayed for EPSPS copy number. Separate qPCR runs to estimate EPSPS gene-copy number were conducted three times on each tissue sample from all cloned plants.
To investigate whether EPSPS copy number is consistent throughout different tissues within an individual plant, five greenhouse-grown plants from the original Georgia population were analyzed, using qPCR to determine EPSPS gene-copy number from six lateral meristems of each plant. Tissue samples were collected from the youngest leaves of each shoot while still connected to the plant, and numbered from 1 (top) to 6 (bottom) to indicate the sampled shoot location within each plant. Three separate qPCR runs to estimate EPSPS gene-copy number were conducted on each tissue sample.
Statistical Analysis
All data were analyzed using R v. 3.0.1 (http://www.r-project.org). Precision and repeatability of EPSPS gene copy-number estimates were assessed by calculating the coefficient of variation (CV) as the ratio of the standard deviation to the mean (Gomez and Gomez Reference Gomez and Gomez2009) for three separate qPCR runs on each of 23 DNA samples. A correlation coefficient was calculated to test the association in F1 progeny between EPSPS gene-copy number and level of glyphosate resistance detected using the shikimate assay. Shapiro-Wilk normality tests were carried out on EPSPS copy numbers for all F1 and F2 families. A linear regression analysis was run between mean EPSPS gene-copy numbers for each parent pair and mean offspring EPSPS gene-copy number (confirmed to be normally distributed via Shapiro-Wilk test) across F1 and F2 populations. A lack-of-fit test performed on the data following the procedures laid out by Kniss and Streibig (Reference Kniss and Streibig2015) confirmed that a linear model fit these data.
Variation in EPSPS copy number among 12 cloned plants after 10 cycles of cloning was examined via a one-way ANOVA, with differences in copy-number estimates from repeated qPCR runs as the error source and using Tukey’s honest significant difference (HSD) for post hoc means separation. To test for gene copy-number differences among shoots on the same plant, a repeated-measures ANOVA was performed on log-transformed data (to meet assumptions of normality) with EPSPS copy number in different shoots on the same plant as the dependent variable. Values of P <0.05 were considered to be statistically significant for all tests.
Results and Discussion
Repeatability of EPSPS Gene-Copy Estimates and Association between Copy Number and Resistance
The CV for three replicated qPCR runs for each of 23 samples ranged from 1.2% to 12.1%, with higher CVs for very high copy-number estimates (>140). The mean CV for the 23 samples was 5.3%, within acceptable limits and indicating that amplified EPSPS gene copy-number estimates based on qPCR in this study were reliable. However, we observed that for 60 of the F1, F2, and cloned plants in this study for which EPSPS copy numbers were assessed in three independently repeated qPCR runs, higher copy-number estimates were associated with increased standard errors (r = 0.612, P <0.001). High EPSPS copy-number estimates based on qPCR should therefore be considered approximate. Shikimate assays on the F1 plants showed that EPSPS gene amplification was consistently associated with glyphosate resistance, with a significant positive correlation between increasing EPSPS copy number and higher levels of glyphosate resistance (r = 0.46, P <0.001).
Inheritance of Amplified EPSPS Gene Copies
To explore the possibility that EPSPS gene amplification is triggered by an allele at a single controlling locus, we grouped F1 and F2 progeny within families based on presence or absence of EPSPS amplification and examined the ratios. Consistent single-gene Mendelian segregation patterns that would have supported this hypothesis were not observed (Table 1).
The association already described between higher EPSPS copy numbers and higher levels of glyphosate resistance suggests that EPSPS amplification may be inherited as a quantitative trait, with additive gene action resulting from independent segregation at amplified EPSPS loci scattered throughout the genome. Eleven F1 families and one F2 family did show normal distributions of EPSPS copy numbers consistent with such a quantitative inheritance pattern, and regression of mean progeny copy numbers for F1 and F2 families on their combined parental copy numbers was significant (Figure 1). Following heritability estimation methods for quantitative traits described by Simmonds (Reference Simmonds1979), we regressed mean offspring copy number on mean (mid-parent) copy number for F1 families and obtained a narrow sense heritability estimate of h 2 = 0.29 (unpublished data). Table 1 shows that having at least one parent with amplified EPSPS copies is generally a predictor of EPSPS amplification in the progeny. However, although most of the F1 families showed a range of EPSPS copy numbers that fell between the two parental copy numbers, copy numbers of full-sib progeny within a family were not clustered around the parent mean as would be expected if amplified EPSPS gene copies are inherited from each parent as independent additive alleles at quantitative trait loci.
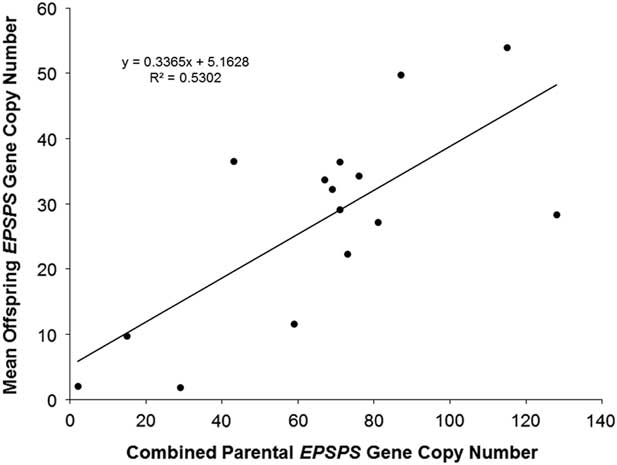
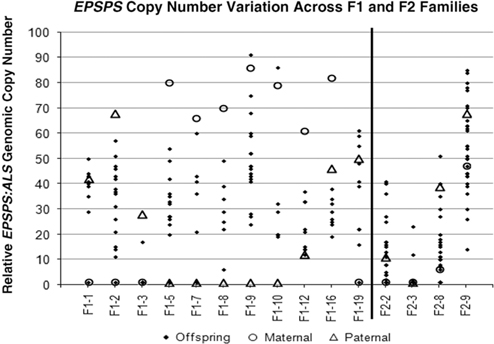
Figure 1 Linear regression of combined parental EPSPS gene-copy number (maternal plus paternal) against mean offspring EPSPS gene-copy number. N = 15, R2 = 0.5302, P-value = 0.002.
Table 1 and Figure 2 show that positive or negative transgressive segregation for EPSPS gene copies occurred in several in F1 families. In F1-3, a cross between an S single-copy female plant and an R male plant with 23 EPSPS gene copies resulted in 17 offspring with one EPSPS gene copy and one offspring with a copy number of 17. For this family, genotyping using ISSR (inter-simple sequence repeat) markers was performed to eliminate the possibility of outside pollen contamination; ISSR markers confirmed that all offspring were true hybrids of the parent plants (unpublished data). Although the paternal parent theoretically contributed multiple EPSPS copies to each offspring, only one of the F1 progeny from this cross had more than a single EPSPS gene. A similar pattern was observed in family F1-16, for which all offspring examined had fewer EPSPS copies than the lower copy-number parent (Table 1; Figure 2). This F1 family showed a distribution of offspring EPSPS copy numbers falling between 19 and 38, despite much higher parental EPSPS copy numbers of 82 and 46 for the maternal and paternal plants, respectively. There is no evidence of maternal or paternal effects on these reduced copy numbers, as the sex of the lower copy-number parent was opposite for these two F1 families. In contrast, F1 families F1-1, F1-9, F1-10, and F1-19 all included at least one offspring with more EPSPS gene copies than the high copy-number parent. As also seen with reduced copy numbers, this occurred equally often with a male or female plant as the high copy-number parent, indicating that gain or loss of gene copies is not subject to maternal or paternal effects. Regression of mean progeny copy numbers on either maternal or paternal F1 or F2 parent copy numbers also showed no significant relationship. These results contradict those of Ribeiro et al. (Reference Ribeiro, Pan, Duke, Nandula, Baldwin, Shaw and Dayan2014), who reported a maternal effect on EPSPS copy number. This is possibly because their A. palmeri populations contained putative apomictic female plants, while we observed no evidence of apomixis.
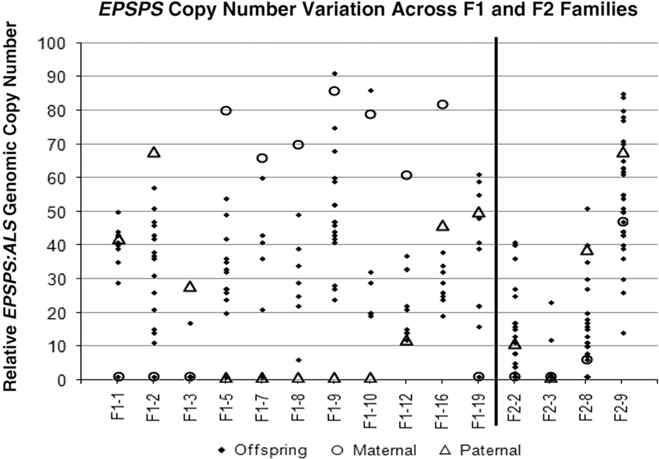
Figure 2 Parental and offspring EPSPS gene-copy numbers for F1 and F2 crosses. Each column represents a single cross or family, consisting a maternal plant (open circle), a paternal plant (triangle), and the progeny (solid diamond).
All four F2 families exhibited transgressive segregation for EPSPS copy number, with both negative and positive transgressive segregation in families F2-8 and F2-9 (Table 1; Figure 2). Three of the four F2 families included offspring for which the EPSPS copy number was greater than the combined copy number of the parents. The most surprising example of this was seen in the F2-3 family, where a cross between two single-copy F1 plants produced 29 plants with a single EPSPS copy, 1 plant with 12 EPSPS copies, and 1 plant with and 23 EPSPS copies. This indicates that within a single generation, progeny from GS low copy-number A. palmeri plants can accumulate sufficient amplified EPSPS copies to produce a GR phenotype, even in the absence of glyphosate. However, we note that the variable EPSPS copy numbers we observed in F1 and F2 progeny should be considered in relation to the within-plant somatic variation for EPSPS copy number that we also observed in this study.
Stability of EPSPS Copy Number among Cloned Generations and within Individual Plants
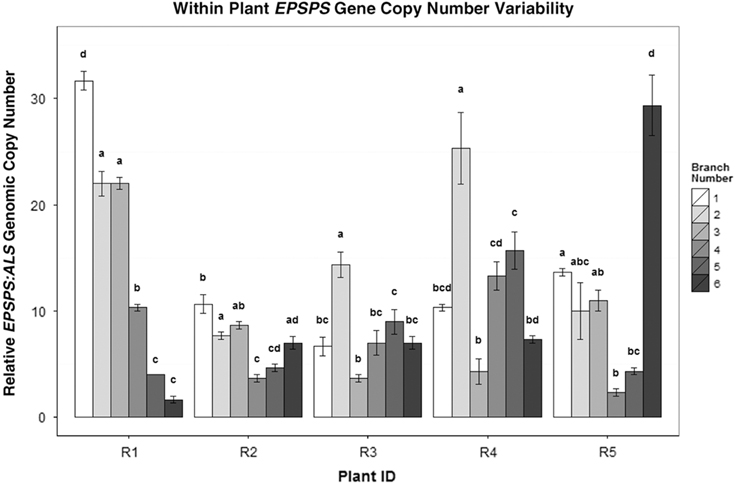
In cloned lines originating from a female A. palmeri plant with 122 EPSPS copies, 10 generations of cloning resulted in plants with EPSPS copy numbers ranging from 40 to 180 (Figure 3). As these plants had never gone through a sexual reproductive phase, any change in gene-copy number presumably occurred in mitotically dividing cells. The same phenomenon was observed in plants in the next (11th) cloned generation (Table 2), revealing further changes in EPSPS copy number after only a single generation of cloning. There were also significant differences in EPSPS copy number among six lateral meristems sampled within all five plants examined from the original Georgia population (F (5, 20) = 10.401, P = 0.01). The largest absolute difference in EPSPS copy number within a single plant ranged from 2 to 32 for plant R1, and plants with higher overall mean EPSPS copy numbers had wider within-plant ranges (Table 2; Figure 4). There was no significant correlation between EPSPS copy number and lateral meristem position on the plant (r = 0.09, n = 30, P = 0.10), and within-plant variation for gene-copy number was found in both male and female plants, suggesting that EPSPS gene amplification was unaffected by physiological factors such as hormonal background or shoot age.
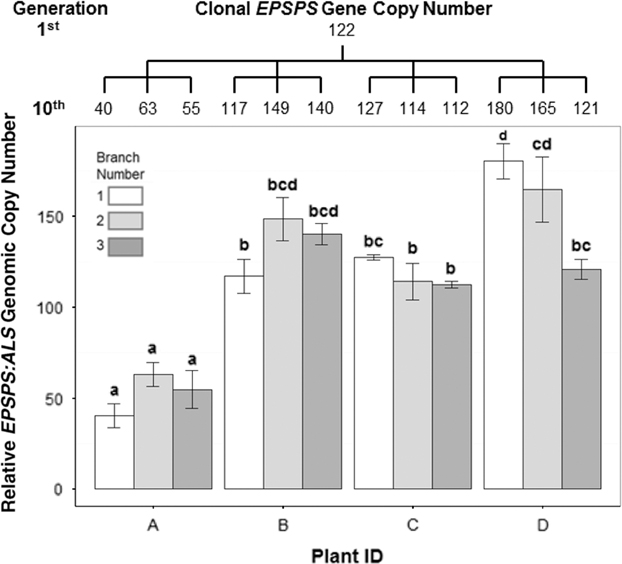
Figure 3 Variation in EPSPS gene-copy number after 10 successive cloned generations starting from a plant with EPSPS copy number = 122. Tree above graph shows mean number of EPSPS gene copies for each clone and relationship between clones. The 10th generation of clones was taken from four plants (A–D), with each plant producing three clones, indicated by the branches of the tree. ANOVA, df = 11, F-value = 22.27, P-value = 5.92e-10. Mean ± SE for each clone with Tukey’s honest significant difference test results shown as letters above columns. Means not sharing the same letter are significantly different.
Table 2 EPSPS gene-copy numbers of Amaranthus palmeri clones derived from a single plant with 122 copies.
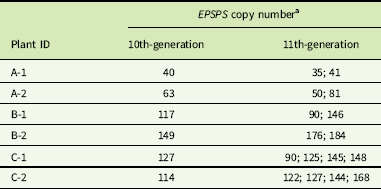
a Gene-copy numbers are shown for 6 individual cloned plants after 10 cycles (generations) of cloning, and for 16 cloned progeny produced from these 6 plants in the next cloning cycle (11th generation).
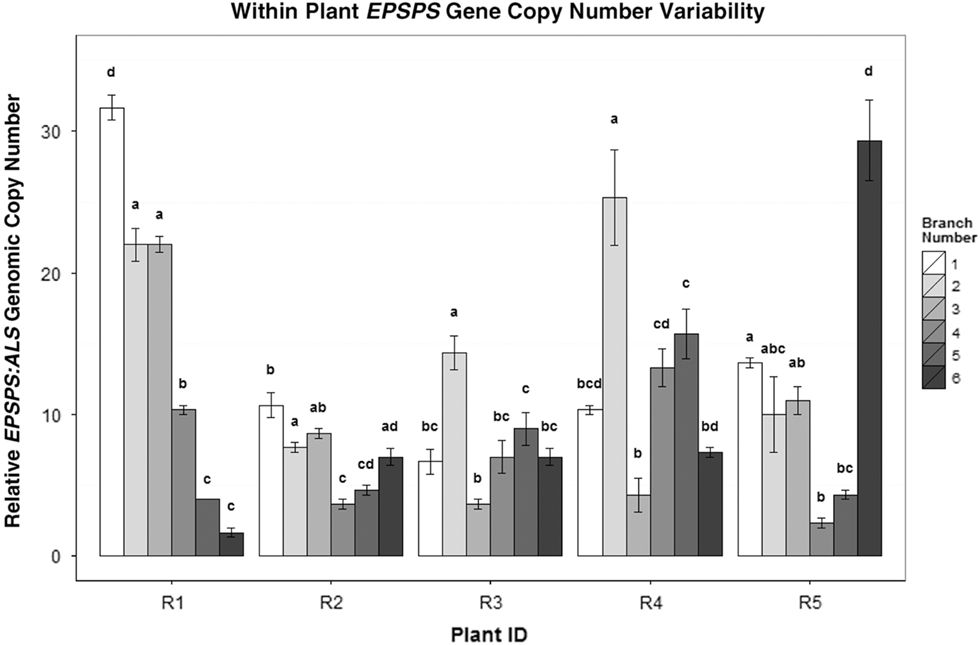
Figure 4 EPSPS copy-number differences between the side shoots of five separate A. palmeri GR plants (R1–R5). Mean ± SE for each clone with Tukey’s honest significant difference (HSD) test results shown as letters above columns. Means not sharing the same letter are significantly different. ANOVAs/Tukey’s HSDs were run on each plant separately to analyze within-plant significant differences in EPSPS gene-copy number.
These results suggest a complex and unstable mechanism in somatic cells is rapidly generating variation in EPSPS copy number, with amplified EPSPS genes being both gained and lost in a single cloning cycle. In a recent paper, Koo et al. (Reference Koo, Molin, Saski, Jiang, Putta, Jugulam, Friebe and Gill2018) reported that amplified EPSPS copies in A. palmeri are located within extrachromosomal circular DNA (eccDNA) molecules tethered to the primary chromosomes and transmitted to daughter cells during mitotic and meiotic cell division. Using FISH (fluorescence in situ hybridization) assays, Koo and coworkers found differing numbers of EPSPS-associated eccDNAs from one cell to the next within the same plant; these eccDNAs appear to replicate autonomously and segregate unequally during the cell cycle. This extrachromosomal distribution of amplified EPSPS gene copies is consistent with the high levels of somatic mosaicism and clonal variation observed in our study and the unpredictable inheritance patterns we found in F1 and F2 families.
Causes and Implications of EPSPS Copy-Number Instability
The transgenerational variability and extensive somatic mosaicism for EPSPS copy number detected in this study have several implications. First, EPSPS copy numbers for individual A. palmeri plants reported in the literature are almost invariably based on one tissue sample, typically a single leaf punch. Our results indicate this may not represent the whole plant, and consequently, the EPSPS copy numbers we report here for F1 and F2 individuals may not reflect the full extent of the variation present. As shown by Koo et al. (Reference Koo, Molin, Saski, Jiang, Putta, Jugulam, Friebe and Gill2018), the number of EPSPS-associated eccDNAs may vary from one cell to the next, meaning any qPCR-based estimate of EPSPS copy number will be an average of the cells in the sample. The most accurate estimate of the EPSPS gene-copy status of an individual A. palmeri plant would ideally be based on the mean and range of EPSPS copy numbers from multiple tissue samples from different parts of the plant.
Second, the positive and negative transgressive segregation seen in our F1 and F2 full-sib families could be explained by the fact that we determined the EPSPS copy number for each parent plant by taking one young leaf from the apical shoot and extracting genomic DNA for qPCR. However, seeds were collected and pooled from the inflorescences on all branches of each female plant. If this maternal parent had varying EPSPS copy numbers in lateral meristematic shoots, progeny germinating from seed produced on these lateral inflorescences could also have different copy numbers. In the same way, varying EPSPS copy numbers in gametophytic tissue in different flowering shoots on a male plant could produce pollen with diverse EPSPS copy numbers. These gene-copy differences among gametes from the male and female parents in a controlled cross could account for the extensive EPSPS copy-number variation we observed within F1 and F2 full-sib families, including gains and losses.
Third, these results show that plants that appear to be fully sensitive to glyphosate may still contain amplifiable EPSPS cassettes. For example, in one of our F2 families (F2-3), we identified increased EPSPS gene-copy numbers in 2 out of 31 full sibs from a cross between two single-copy F1 parents that were both confirmed as GS via shikimate assay. In the context of eccDNA-based EPSPS gene amplification, one or both of these parent plants may have a low frequency of cells with eccDNAs containing EPSPS copies that were transmitted during meiosis to the gametophyte. Koo et al. (Reference Koo, Molin, Saski, Jiang, Putta, Jugulam, Friebe and Gill2018) also observed higher numbers of eccDNAs in the actively dividing cells of an F1 plant (mitotic root tip and meiotic cells) than in the leaf somatic cells, providing a possible explanation for the results seen here. Whether meristematic tissue in general has higher overall EPSPS gene-copy number than mature leaf tissue is unknown and requires further investigation.
The implications of this finding are troublesome: although a sampled leaf may return an average EPSPS copy number of one, the vagaries of eccDNA behavior during cell division could result in a rapid increase in copy number under selection pressure from glyphosate application. This form of “hidden resistance” not only makes diagnosing glyphosate resistance in the field problematic, but also highlights the unusual nature of this adaptation in A. palmeri. By maintaining amplified EPSPS copies in the extrachromosomal gene space, these plants retain the capacity for rapid increases or decreases in the number of EPSPS genes, potentially in a single generation. In addition, the lack of fitness costs associated with increased EPSPS gene copies in GR A. palmeri (Giacomini et al. Reference Giacomini, Westra and Ward2014; Vila-Aiub et al. Reference Vila-Aiub, Goh, Gaines, Han, Busi, Yu and Powles2014) means this resistance mechanism is likely to persist and spread.
Fourth, based on the clonal results in this study, EPSPS copy-number variation appears to vary randomly among plant cells, possibly resulting from mis-segregation of eccDNAs with each round of cell division (Koo et al Reference Koo, Molin, Saski, Jiang, Putta, Jugulam, Friebe and Gill2018). Stress-induced gene amplification has been reported in Escherichia coli (Slack et al. Reference Slack, Thornton, Magner, Rosenberg and Hastings2006). However, the plants in our study were all grown under optimal greenhouse conditions in the absence of glyphosate, so it is unlikely that EPSPS gene amplification occurring in the course of this study was induced by abiotic stress. It is more likely that the random segregation of eccDNAs containing EPSPS genes led to some cells with many copies and some with few or none. How these gene copies are produced is still unknown, although a mechanism driven by transposon-mediated excision and replication of the EPSPS cassette via an extrachromosomal rolling-circle mechanism has been suggested as the basis for EPSPS amplification in A. palmeri (Molin et al. Reference Molin, Wright, Lawton-Rauh and Saski2017). Furthermore, this cassette was also found to contain an autonomous replication sequence, similar to those found in self-replicating yeast eccDNAs (Møller et al. Reference Møller, Parsons, Jørgensen, Botstein and Regenberg2015), providing a mechanism for replication of the EPSPS copies once they are moved into the extrachromosomal space. This ability to self-replicate benefits the plant by allowing faster evolutionary adaptation in response to stress. Whether other stress-adaptive genes are also present in extrachromosomal DNA is currently unexplored.
To summarize: inheritance of EPSPS-amplification-based glyphosate resistance in A. palmeri follows a non-Mendelian pattern of inheritance with both positive and negative transgressive segregation for EPSPS copy number. These complex and unpredictable inheritance patterns appear to be the result of extensive somatic mosaicism for EPSPS copy number within individual plants, and of variable transmission of EPSPS copies from one generation to the next via sexual and asexual reproduction. In the most extreme examples, EPSPS copy number was found to vary by an order of magnitude within a plant, highlighting the need to sample multiple tissues from a single plant when assaying for resistance. This somatic mosaicism and unpredictable transgenerational inheritance is consistent with the presence of the amplifiable EPSPS cassette on eccDNA, tethered to the primary chromosomes but inherited separately and thus allowing for mis-segregation of gene copies during mitosis and meiosis. The presence of high EPSPS copy-number plants in offspring from a cross between two GS parents demonstrates how even if only a small percentage of the cells in a plant retain an elevated number of EPSPS copies, these may be transmitted to the next generation. This genomic plasticity allows A. palmeri to respond rapidly to its environment, giving the plant a high level of adaptive flexibility.
Acknowledgments
We would like to thank Doug Sammons for providing advice and guidance on this work as well as for contributing clonal populations for study. Support for this research was provided by Monsanto. No conflicts of interest have been declared.