Palmer amaranth is ranked as the most troublesome and one of the most common weeds found in North America (Wychen Reference Wychen2016). Palmer amaranth is especially prevalent in the midsouthern United States, and its control is a major concern of soybean and cotton growers and consultants in these states (Prostko Reference Prostko2011; Riar et al. Reference Riar, Norsworthy, Steckel, Stephenson, Eubank and Scott2013). Control of Palmer amaranth has become increasingly difficult over the past decade because of the widespread distribution of biotypes resistant to acetolactate synthase (ALS) inhibitors and glyphosate in the South (Bond et al. Reference Bond, Oliver and Stephenson2006; Heap Reference Heap2017). As a consequence, growers have become increasingly dependent on PRE and POST applications of protoporphyrinogen oxidase (PPO) inhibitors, and this appears to be a commonly adopted weed management tactic for control of ALS inhibitor–resistant and glyphosate-resistant broadleaf weeds in soybean and cotton (Owen and Zelaya Reference Owen and Zelaya2005; Rousonelos et al. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012).
PPO-inhibiting herbicides prevent the conversion of protoporphyrinogen IX to protoporphyrin IX by the plastid-localized PPO enzyme (PPX2) (Matringe et al. Reference Matringe, Camadro and Scalla1989; Jacobs and Jacobs Reference Jacobs and Jacobs1993). PPO inhibitor–resistant Palmer amaranth was first discovered in 2011 in Arkansas and then in Tennessee and Illinois in 2015 and 2016, respectively (Heap Reference Heap2017). Initially, the mechanism of resistance to PPO inhibitors in Palmer amaranth was documented as being the same as in common waterhemp (Amaranthus rudis Sauer) (Salas et al. Reference Salas, Burgos, Tranel, Singh, Glasgow, Scott and Nichols2016; Thinglum et al. Reference Thinglum, Riggins, Davis, Bradley, Al-Khatib and Tranel2011). The target-site resistance mechanism involves a codon deletion in the PPX2 gene, resulting in the loss of a glycine residue at the 210th position (ΔG210) of the PPO enzyme (Lee et al. Reference Lee, Hager and Tranel2008; Patzoldt et al. Reference Patzoldt, Hager, McCormick and Tranel2006). Giacomini et al. (Reference Giacomini, Umphres-Lopez, Nie, Mueller, Steckel, Young and Tranel2017) reported two new mutations encoding for a glycine (G) or a methionine (M) instead of an arginine at the 128th (R128) amino acid residue (referred to as R98 in their study) in the PPO enzyme of Palmer amaranth. The R128 amino acid is homologous to the R98 in common ragweed (Ambrosia artemisiifolia L.), in which an R98 to leucine amino acid change was shown to confer resistance to fomesafen (Rousonelos et al. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012; Salas et al. Reference Salas, Burgos, Rangani, Singh, Refatti, Piveta, Tranel, Mauromoustakos and Scott2017). The difference in the amino acid number is due to the signal peptide (30 aa in length) found in the PPX2 protein of Amaranthus spp. but not in common ragweed (Dayan et al. Reference Dayan, Daga, Duke, Lee, Tranel and Doerksen2010; Rousonelos et al. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012).
The R128G/M mutations in Palmer amaranth were identified in an accession from Woodruff County in Arkansas and in two accessions from Lauderdale and Shelby counties in Tennessee (Giacomini et al. Reference Giacomini, Umphres-Lopez, Nie, Mueller, Steckel, Young and Tranel2017). Interestingly, the Woodruff County accession was segregating for the ΔG210 deletion and the R128G mutation, and this accession was shown to exhibit cross-resistance to the PPO-inhibiting herbicides fomesafen (diphenylethers), flumioxazin (N-phenylphthalimide), oxadizon (oxadiazole), sulfentrazone (triazolinone), and saflufenacil (pyrimidindione), and presumably to other herbicides from their respective chemical families (Schwartz-Lazaro et al. Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017).
A 2016 survey of soybean consultants from Arkansas, Louisiana, southeast Missouri, Mississippi, and Tennessee found 79% of the hectares received multiple applications of PPO inhibitors in the past 3 yr, and 69% of the consultants suspected PPO inhibitor–resistant Palmer amaranth in at least one of their scouted fields (JK Norsworthy, personal observations). Thus, a large-scale screen of 227 Palmer amaranth accessions collected from the major row crop–producing counties (29 counties) in Arkansas was conducted to determine the severity of PPO-inhibitor resistance in Arkansas and to further identify the dominant resistance mechanism(s) in these accessions.
Materials and Methods
Plant Material
A late-season weed-escape survey was conducted in the fall of 2016. Inflorescences from Palmer amaranth accessions were collected from crop production fields (mainly soybean) across the state of Arkansas by our research group, growers, consultants, or extension agents and sent to the Altheimer Laboratory at the University of Arkansas, Fayetteville, AR. The GPS coordinates for all accessions were recorded. In total, 227 accessions were screened for PPO-inhibitor resistance, and the seed lot for each accession, on average, was created by threshing 10 inflorescences.
PPO-Inhibitor Resistance Screen
In the greenhouse, each accession was screened for PPO-inhibitor resistance against a rate of fomesafen that completely controlled a susceptible standard. Seeds from each accession were germinated in 50-well plastic trays filled with potting mix (Sunshine® Premix No. 1, Sun Gro Horticulture, Bellevue, WA). One week after germination, seedlings were thinned to 1 plant well−1. Plants were grown under a 16-h photoperiod and a 35/25 C day/night temperature. Once seedlings reached a 5- to 7-cm height (3- to 4-leaf stage), they were sprayed with fomesafen at 395 g ai ha−1 (Flexstar® 1.88 EC, Syngenta, Greensboro, NC) using a research track sprayer equipped with two flat-fan spray nozzles (TeeJet® spray nozzles, Spraying System, Wheaton, IL) calibrated to deliver 187 L ha−1 of herbicide solution at 269 kPa, moving at 1.6 km h−1. A nonionic surfactant was included at 0.25% v/v. At 21 d after treatment (DAT), dead or alive counts were recorded for each accession and expressed as percent mortality. The experiment was conducted in two runs in a completely randomized design with 1 replication run−1 (50 plants run−1). All maps presented in this paper were made using the ggplot in R software (R Core Team 2017).
TaqMan® qPCR Assay for Detection of the ΔG210 and R128G/M Mutations
After percent mortality was recorded, young leaf tissue was harvested from a maximum of 20 plants from each accession that had survivors. In total, 167 accessions were genotypically screened out of the original 227 accessions in the greenhouse screen. Harvested tissue from each plant was placed into separate 1.5-mL microfuge tubes (Thermo Fisher Scientific, Waltham, MA) and subsequently stored at −80 C until use. Genomic DNA (gDNA) was isolated from each plant sample using a modified CTAB (cetyl trimethylammonium bromide) protocol (Doyle and Doyle Reference Doyle and Doyle1987). The TaqMan® quantitative polymerase chain reaction (qPCR) allelic discrimination (AD) assay was performed using a CFX 96 real-time detection system (Bio-Rad, Hercules, CA) to detect the presence/absence of the ΔG210 codon deletion or the R128G or R128M mutation in the PPX2 gene of collected plants. Fluorescent probes (6-carboxyfluorescein, FAM and VIC labeled) were used to discriminate between the resistant and susceptible alleles of the PPX2 gene in the accessions. For each assay, the qPCR reaction mix (10 µl) consisted of 2 µl of GoTaq® Flexi buffer (Promega, Madison, WI), 1.2 µl of 25 mM MgCl2 (Promega), 0.5 µl of 10 mM dNTP mix (Promega), 0.5 µl of primer-probe mix (custom TaqMan® SNP genotyping assay, Thermo Fisher Scientific), 0.1 µl GoTaq® Flexi DNA polymerase (Promega), 1 µl of gDNA, 4.7 µl of molecular grade water. The qPCR conditions were 95 C for 3 min, 40 cycles of 95 C for 15 s, and an annealing at 60 C for 1 min followed by a plate read on every cycle. All plates included a homozygous susceptible (1986) and a heterozygous resistant plant (ΔG210) as controls. The Bio-Rad CFX software was used to analyze the qPCR allelic discrimination data expressed in relative fluorescence units. The TaqMan primer and probe combinations used for detection of the ΔG210 codon were previously reported in Giacomini et al. (Reference Giacomini, Umphres-Lopez, Nie, Mueller, Steckel, Young and Tranel2017). Detection of the ΔG210 codon deletion was done for each harvested plant to calculate the resistance frequency within an accession. The resistance frequency (%) was calculated using the following equation:
 $$\eqalignno{ {\rm Frequency}&\,{\equals}\left[ {{{\left( {\displaystyle{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm homozygous}\,{\rm for}\,\Delta {\rm G}210{\plus}\atop\displaystyle{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm heterozygous}\,{\rm for}\,\Delta {\rm G}210} \right)} \over {{\rm total}\,{\rm number}\,{\rm of}\,{\rm plants}\,{\rm used}\,{\rm for}\,{\rm AD}\,{\rm assay}}}} \right]\cr&\quad\times\,\%\,\,{\rm survival}\,{\rm in}\,{\rm the}\,{\rm greenhouse}$$
$$\eqalignno{ {\rm Frequency}&\,{\equals}\left[ {{{\left( {\displaystyle{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm homozygous}\,{\rm for}\,\Delta {\rm G}210{\plus}\atop\displaystyle{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm heterozygous}\,{\rm for}\,\Delta {\rm G}210} \right)} \over {{\rm total}\,{\rm number}\,{\rm of}\,{\rm plants}\,{\rm used}\,{\rm for}\,{\rm AD}\,{\rm assay}}}} \right]\cr&\quad\times\,\%\,\,{\rm survival}\,{\rm in}\,{\rm the}\,{\rm greenhouse}$$
In contrast, gDNA was pooled from up to 10 individual plants within an accession to perform a quick assay for detecting the presence/absence of mutations at the 128th amino acid position of PPX2 gene in each accession. Common forward (5′-TCTTCTGTCACAGCCAATTTCACA-3′) and reverse (5′-CAGAGGACTTACTAGCACAGGAAGA-3′) primers were designed to amplify the regions flanking the coding region of the 128th amino acid. The probes used for the R128G assay were 5′-AAATAAAAGGTACATAGCTAG-3′ and 5′-AAATAAAGGGTACATAGCTA-3′, whereas probes used for the R128M assay were: 5′-ATAAAAGGTACATAGCTAGAG-3′ and 5′-ATAAAATGTACATAGCTAG-3′. The nucleotides that are underlined in the probe sequences indicate wild-type (AGG) and mutated (GGG or ATG) alleles at the R128 region of the PPX2 gene.
Results and Discussion
Sensitivity of Arkansas Palmer Amaranth Accessions to Fomesafen
To characterize the efficacy of PPO inhibitors in Arkansas, the progeny from 227 accessions were sprayed with a labeled rate of fomesafen (395 g ha−1). Accessions were grouped based on whether mortality was ≥90% or <90% at 21 DAT (Figure 1; Table 1). The 90% mortality threshold was chosen based on a previously conducted field survey of soybean farmers in which they indicated >90% mortality as an acceptable level of weed control for POST herbicides (Norsworthy Reference Norsworthy2003). A known susceptible accession (1986) was included in the screen as a control and, as expected, had 100% mortality at 21 DAT (unpublished data). Out of 227 accessions, 108 had ≥90% mortality, with 44 accessions having no survivors at 21 DAT (Table 1). In contrast, 52% of the screened accessions had <90% mortality, and these accessions could be found in 21 out of the 29 counties sampled in this study (Figure 1). For example, 24 out of the 39 accessions collected from Crittenden County had <90% mortality, indicating growers in this county should assume PPO-inhibiting herbicides will not adequately control Palmer amaranth. In general, accessions located in the northeastern and mid-eastern (above 35.5°N) parts of Arkansas were poorly controlled (<90% mortality), indicating that gene flow of putative fomesafen resistance alleles is widespread and troublesome in the aforementioned regions.
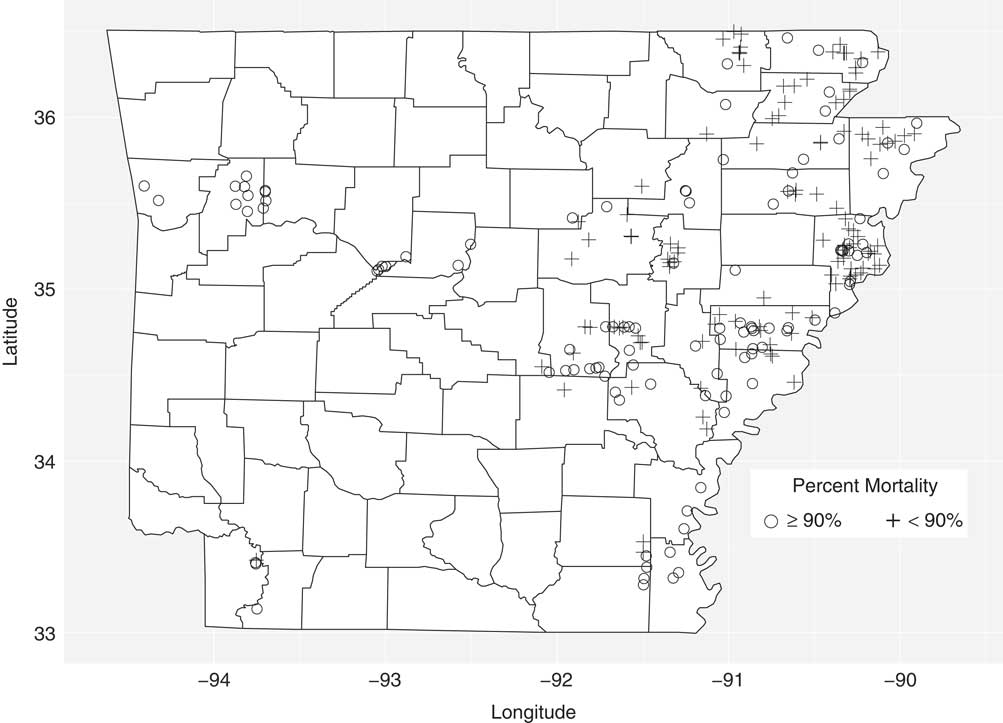
Figure 1 The susceptibility of 227 Palmer amaranth accessions collected from the major row crop–producing areas in Arkansas to fomesafen at 395 g ha−1. At 21 d after treatment, dead/alive counts were converted to percent mortality, and accessions were grouped based on whether mortality was ≥90% or <90% with fomesafen.
Table 1 Palmer amaranth accessions treated with 395 g ha−1 of fomesafen in the greenhouse and screened for PPO-inhibitor resistance.Footnote a
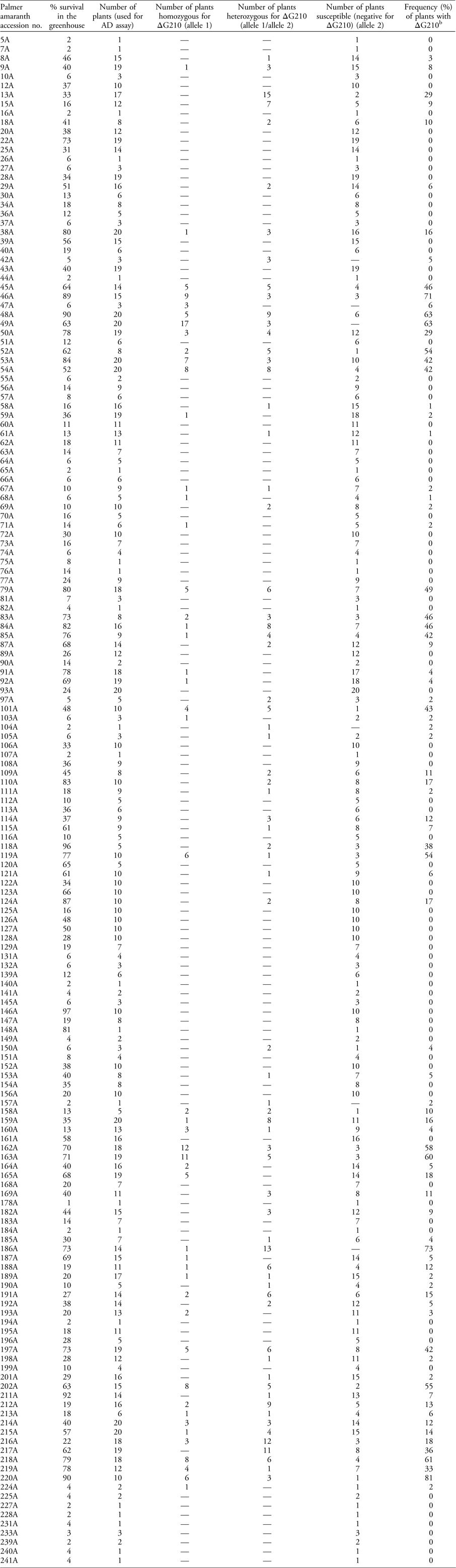
a Percent survival was calculated out of 100 treated plants. The survivors of the herbicide treatment were subjected to an allelic discrimination (AD) qPCR assay to detect the presence/absence of the ΔG210 codon deletion in the PPX2 gene and its corresponding frequency in each accession. The number of homozygous and heterozygous individuals in each accession with respect to the ΔG210 deletion conferring PPO—inhibitor resistance is also shown. A dash (—) indicates zero plants were found to contain this genotype.
b
![]() $${\rm Frequency}{\equals}\left[ {{\displaystyle{\left( {{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm homozygous}\,{\rm for}\,\Delta {\rm G}210{\plus}{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm heterozygous}\,{\rm for}\,\Delta {\rm G}210} \right)} \over {\displaystyle{\rm total}\,{\rm number}\,{\rm of}\,{\rm plants}\,{\rm used}\,{\rm for}\,{\rm AD}\,{\rm assay}}}} \right]{\times}\,\%\,\,{\rm survival}\,{\rm in}\,{\rm the}\,{\rm greenhouse}$$
$${\rm Frequency}{\equals}\left[ {{\displaystyle{\left( {{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm homozygous}\,{\rm for}\,\Delta {\rm G}210{\plus}{\rm no}{\rm .}\,{\rm of}\,{\rm plants}\,{\rm heterozygous}\,{\rm for}\,\Delta {\rm G}210} \right)} \over {\displaystyle{\rm total}\,{\rm number}\,{\rm of}\,{\rm plants}\,{\rm used}\,{\rm for}\,{\rm AD}\,{\rm assay}}}} \right]{\times}\,\%\,\,{\rm survival}\,{\rm in}\,{\rm the}\,{\rm greenhouse}$$
Identification and Distribution of PPO-Inhibitor Resistance Mechanisms
To confirm resistance and identify the underlying mechanism(s) of resistance to fomesafen in the Arkansas accessions, a TaqMan AD assay was conducted on the surviving plants in the greenhouse screen to detect the presence/absence of the ΔG210 and R128G/M mutations in the target-site PPX2 gene (Table 2). These mechanisms are known to confer resistance to PPO-inhibiting herbicides and have been previously reported to exist in a Palmer amaranth accession from Arkansas or in the neighboring state of Tennessee (Giacomini et al. Reference Giacomini, Umphres-Lopez, Nie, Mueller, Steckel, Young and Tranel2017; Salas et al. Reference Salas, Burgos, Rangani, Singh, Refatti, Piveta, Tranel, Mauromoustakos and Scott2017). Out of 227 total accessions screened, only 183 accessions had plants alive at 21 DAT; however, a number (16) of accessions had one or two survivors with insufficient tissue for gDNA isolation and therefore were not further analyzed (Table 1). Thus, the TaqMan assay was run on 167 out of the total 227 accessions (Table 2). Several samples were further validated by Sanger DNA sequencing (Genewiz, South Plainfield, NJ), which revealed a high reliability of the TaqMan assay method for screening mutations in the herbicide target-site regions in various accessions (unpublished data).
Table 2 The presence (+) or absence (−) of target-site deletion (ΔG210) and mutations (R128G/M) in the 167 Palmer amaranth accessions from Arkansas.a

a A TaqMan qPCR assay was conducted to screen for mutation(s) conferring PPO-inhibitor resistance in Palmer amaranth. Out of the 227 total accessions screened in the greenhouse, only 183 accessions had plants alive at 21 d after fomesafen treatment and a number (16) of accessions had only one or two survivors.
Distribution of the ΔG210 Resistance Allele
Of the 167 Palmer amaranth accessions that were genotypically analyzed in this study, only 109 accessions were found to harbor a known PPO inhibitor–resistant allelic form of the PPX2 gene (Figure 2; Table 2). PPO inhibitor–resistant Palmer amaranth was found in 18 out of the 29 major agriculture-producing counties in Arkansas, and the ΔG210 resistance mechanism was detected in at least one accession from each of those 18 counties (Figure 2). Genotypically, 49% of the analyzed accessions had at least one plant that harbored an allele of the PPX2 gene encoding a ΔG210 deletion. The frequency of plants with a ΔG210 allele (combined over homozygous and heterozygous plants) ranged from 1% to 81%, and on average was 20% (Table 1). However, this underestimates the overall frequency of resistant plants, because 27% of the accessions were found to be segregating for both ΔG210 and R128G/M mutations (Table 2). The majority (53%) of accessions harboring the ΔG210 mechanism were located in the northeastern part of the state surrounding the Missouri bootheel (specifically Clay, Greene, Randolph, Lawrence, Craighead, and Mississippi counties) (Figure 2).
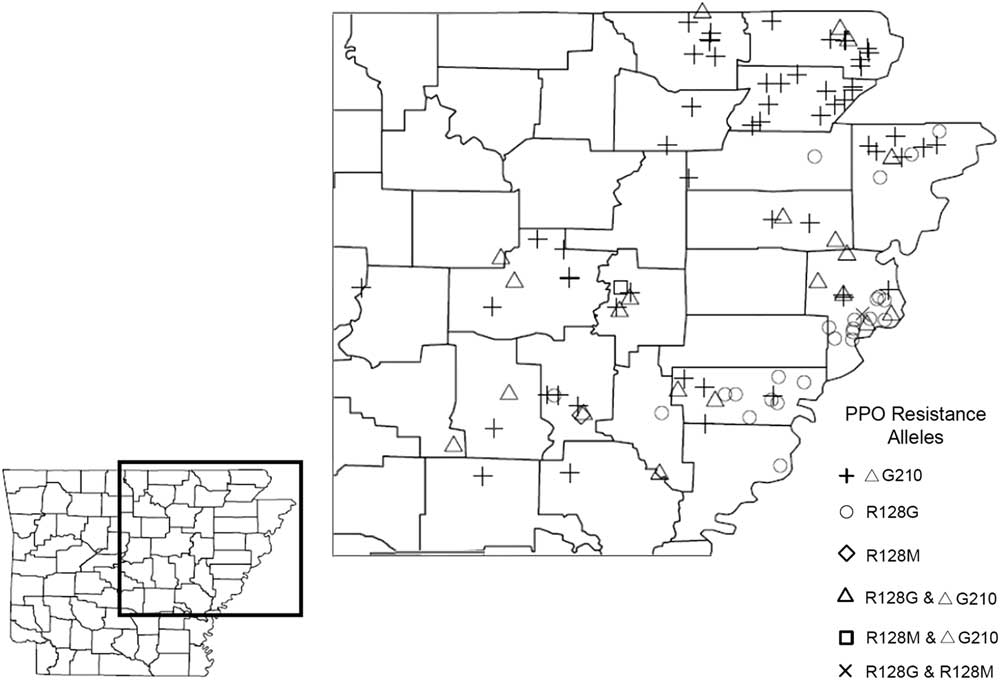
Figure 2 The confirmation and distribution of PPO-inhibitor resistance alleles in Palmer amaranth accessions from Arkansas. A TaqMan qPCR allelic discrimination assay was used to detect the presence or absence of expected target-site resistance mechanisms (ΔG210 codon deletion, R128G, and R128M) in the PPX2 gene of Palmer amaranth.
Distribution of the R128G and R128M Resistance Alleles
The R128G allele was discovered in 28% of the 167 accessions screened and found in 12 out of the 29 major agriculture-producing counties in Arkansas (Figure 2; Table 2). Nearly 55% of the accessions harboring the R128G allele were found in Crittenden and Lee counties near Memphis, TN. The R128M allele, on the other hand, was rare and was found only in one accession each from Prairie (12A), Woodruff (87A), and Crittenden (147A) counties. The large spatial separation of the R128M allele is most likely the result of seed-mediated gene flow or occurred through independent evolutionary events (Jasieniuk et al. Reference Jasieniuk, Brule-Babel and Morrison1996). The frequency of the R128G or R128M allele was not determined, because individuals were pooled within each accession to rapidly screen for the presence/absence of these mutations.
Additionally, 27 out of the 167 accessions genetically screened in this study were not adequately controlled (<90% mortality) with fomesafen and did not contain any known resistant alleles (ΔG210 or R128G/M) (Tables 1 and 2). The surviving plants within these accessions exhibited moderate to severe injury; however, by 21 DAT, regrowth was observed from either the apical or the lateral meristems. These accessions are scattered throughout the Arkansas growing region (unpublished data). This may indicate the presence of a novel target-site or non−target site PPO-inhibitor resistance mechanism(s) (such as herbicide metabolism) existing in Arkansas. These 27 accessions need to be investigated further for unknown resistance mechanism(s).
In this study, PPO inhibitor–resistant Palmer amaranth was found in 18 out of the 29 major agriculture-producing counties in Arkansas. This strongly indicates that PPO-inhibitor resistance in Palmer amaranth is widespread in Arkansas, with the most common resistance-conferring alleles being ΔG210 and R128G (Figure 2; Table 2). These resistance mechanisms have been shown to confer cross-resistance to the PPO-inhibitor chemical families: diphenylethers, N-phenylphthalimide, oxadiazole, triazolinone, and pyrimidindione (Rousonelos et al. Reference Rousonelos, Lee, Moreira, VanGessel and Tranel2012; Schwartz-Lazaro et al. Reference Schwartz-Lazaro, Norsworthy, Scott and Barber2017), indicating growers cannot rely on PPO-inhibiting herbicides to control these biotypes. In fields infested with PPO inhibitor–resistant and glyphosate-resistant Palmer amaranth, growers will need to rotate crops; apply herbicides with different sites of action (e.g., WSSA Groups 5 and 15); use herbicide-resistant trait technologies (LibertyLink®, Xtend®, or Enlist®); and incorporate cultural techniques such as cover crops, tillage, or hand-weeding.
Acknowledgments
Funding for this research was provided by the Arkansas Soybean Promotion Board.






