Article contents
Higher-Order Structure of Human Chromosomes Observed by Electron Diffraction and Electron Tomography
Published online by Cambridge University Press: 20 November 2020
Abstract
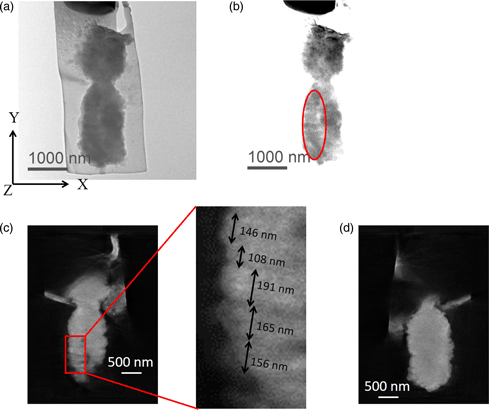
It is well known that two DNA molecules are wrapped around histone octamers and folded together to form a single chromosome. However, the nucleosome fiber folding within a chromosome remains an enigma, and the higher-order structure of chromosomes also is not understood. In this study, we employed electron diffraction which provides a noninvasive analysis to characterize the internal structure of chromosomes. The results revealed the presence of structures with 100–200 nm periodic features directionally perpendicular to the chromosome axis in unlabeled isolated human chromosomes. We also visualized the 100–200 nm periodic features perpendicular to the chromosome axis in an isolated chromosome whose DNA molecules were specifically labeled with OsO4 using electron tomography in 300 keV and 1 MeV transmission electron microscopes.
- Type
- Biological Applications
- Information
- Copyright
- Copyright © The Author(s), 2020. Published by Cambridge University Press on behalf of the Microscopy Society of America
Footnotes
Equally contributed to this work.
References
- 5
- Cited by





