Article contents
The comparison of standard and fully recursive multivariate models for genetic evaluation of growth traits in Markhoz goat: predictive ability of models and ranking of animals
Published online by Cambridge University Press: 23 October 2020
Abstract
Data of 2780 Markhoz kids originated from 1216 dams and 211 sires during 1993–2016 in Markhoz Goat Breeding Station, located in Sanandaj, Iran, were used. Traits investigated were body weights at birth, weaning, six-month age [six months weight (6MW)], nine-month age and yearling age [yearling weight (YW)]. Two considered multivariate models including standard multivariate model (SMM) and fully recursive multivariate model (FRM) were compared using deviance information criterion (DIC) and predictive ability measures including mean square of error (MSE) and Pearson's correlation coefficient between the observed and predicted values $(r(y,\hat{y}))$ of records. Spearman's rank correlation coefficients between posterior means of direct genetic effects of the studied traits of kids under SMM and FRM were also calculated across all, 50, 10 and 1% top-ranked animals. In general, FRM performed better than SMM in terms of lower DIC and MSE and also higher $r\lpar y\comma \;\hat{y}\rpar$
of records. Spearman's rank correlation coefficients between posterior means of direct genetic effects of the studied traits of kids under SMM and FRM were also calculated across all, 50, 10 and 1% top-ranked animals. In general, FRM performed better than SMM in terms of lower DIC and MSE and also higher $r\lpar y\comma \;\hat{y}\rpar$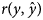 . For all traits, the lowest MSE and the highest $r\lpar y\comma \;\hat{y}\rpar$
. For all traits, the lowest MSE and the highest $r\lpar y\comma \;\hat{y}\rpar$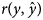 were obtained under FRM. All structural coefficients estimated under FRM were statistically significant except for that of 6MW on YW. Comparisons of Spearman's rank correlations between posterior means of direct genetic effects of kids for growth traits under SMM and FRM revealed that taking the causal relationships among the studied growth traits of Markhoz goat into account may cause considerable re-ranking for the animals in terms of estimated breeding values, especially for the top-ranked animals. It may be concluded that FRM had more plausibility over SMM for genetic evaluation of the studied growth traits in Markhoz goat.
were obtained under FRM. All structural coefficients estimated under FRM were statistically significant except for that of 6MW on YW. Comparisons of Spearman's rank correlations between posterior means of direct genetic effects of kids for growth traits under SMM and FRM revealed that taking the causal relationships among the studied growth traits of Markhoz goat into account may cause considerable re-ranking for the animals in terms of estimated breeding values, especially for the top-ranked animals. It may be concluded that FRM had more plausibility over SMM for genetic evaluation of the studied growth traits in Markhoz goat.
- Type
- Animal Research Paper
- Information
- Copyright
- Copyright © The Author(s), 2020. Published by Cambridge University Press
References
- 1
- Cited by





