INTRODUCTION
The cell wall-less bacterium Mycoplasma pneumoniae is an important pathogen of the human respiratory tract causing a range of manifestations, e.g. pharyngitis, bronchitis and tracheobronchitis, to cases of primary atypical, interstitial pneumonia [Reference Fernald, Collier and Clyde1, Reference Clyde, Tully and Whitcomb2]. The clinical signs of M. pneumoniae-induced pneumonia are usually characterized by a non-specific and slowly progressing course of the infection with non-productive cough and moderate fever. Recent studies demonstrated that M. pneumoniae is responsible for 5–30% of all cases of community-acquired pneumonia [Reference Blasi3–Reference Huang9]. Despite clinical resolution after adequate antibiotic therapy patients with respiratory infections due M. pneumoniae may continue to shed the bacteria [Reference Smith, Friedewald and Chanock10]. Furthermore, the primary infections of the upper or lower respiratory tract can be followed by extrapulmonary complications [Reference Tsiodras11]. The incidence of M. pneumoniae infections is strongly influenced by the detection method used in the study, the age structure of the patient group investigated and the time-dependent occurrence of infections. It is well known that epidemic peaks of diseases caused by M. pneumoniae occur at time intervals of 3–7 years [Reference Lind12, Reference Eun13] whereas endemic outbreaks in closed populations such as in schools or in army camps are reported more frequently [Reference Waites and Talkington14].
Due to the fact that cultivation methods are laborious, insensitive and time-consuming, routine diagnosis of the M. pneumoniae infections are mostly based on serological and PCR approaches. The practical significance of serological tests depends on the time of serum sampling during the course of the disease, the age of the patient, the occurrence of significant levels of specific (mainly IgG) antibodies due to previous infections with M. pneumoniae, the availability of paired sera, and also on the test kit used for diagnosis [Reference Daxboeck, Krause and Wenisch15]. Because of the high sensitivity and specificity of molecular methods many of the limitations of the classical test procedures for detecting M. pneumoniae infections have been overcome [Reference Loens, Ursi, Goossens and Ieven16].
Previous investigations found that M. pneumoniae clinical isolates can be divided into different subtypes and variants showing sequence variations in proteins of the adhesion complex [Reference Jacobs17]. These genotypes tend to dominate in a time-dependent frequency with subtype changes in the follow-up of epidemic peaks, suggesting a possible role of the type distribution in the epidemiology of infections caused by M. pneumoniae [Reference Kenri18, Reference Dumke19]. Up to now, investigations of the circulation of different subtypes and variants in human populations and their role in the incidence of infections have been performed by molecular characterization of isolated strains [Reference Dumke19–Reference Schwartz21] and specimens of M. pneumoniae-positive patients [Reference Dumke22]. The strains differ in the sequence of the two repetitive elements (repMp) located in the main P1 adhesin. In addition to this ‘antigen-based’ subtyping approach, it can be assumed that the quantitative detection of antibodies directed against subtype- and variant-specific protein regions of the main immunogens of M. pneumoniae in sera of infected persons also leads indirectly to genotyping results. Thus, the host's immune response to the actual strain might be used for ‘antibody-based’ epidemiological subtyping [Reference Dumke, Schurwanz and Jacobs23].
In order to investigate the incidence and the different aetiological agents of non-nosocomial pneumonia within adult outpatients, the German network of competence for community-acquired pneumonia (CAPNETZ; http://www.capnetz.de) was established in 2003. Organization and data collection within the network have been described previously [Reference Welte, Suttorp and Marre24]. Briefly, the network included local clinical centres (LCC) coordinating the data and sample collection in a number of associated general medical practices. Local centres were selected by their pulmonological competence. Preparation of the serum and respiratory tract samples and organization of the sample transport to the different reference laboratories were carried out by a specialized centre. In the current study we summarize the results of molecular subtyping of M. pneumoniae-positive respiratory tract specimens and the local distribution of the different subtypes and variants of M. pneumoniae. Besides the direct PCR-based subtyping we present for the first time results that correlate the occurrence of subtype- and variant-specific antibodies in the sera of M. pneumoniae-positive pneumonia patients with the genotype of M. pneumoniae causing this disease. Since sera are often collected from patients with symptoms of respiratory tract infections this approach offers the opportunity to investigate large numbers of samples in order to contribute to the understanding of the epidemiology of M. pneumoniae infections.
METHODS
Patients and samples
Samples were collected during the period 2003–2006 within the CAPNETZ network (Fig. 1). The criteria for inclusion of outpatients in the CAPNETZ study were age ⩾18 years, the presence of a new infiltrate on chest radiography, and at least one of the following criteria: history of fever (temperature ⩾38·3°C), cough, production of sputum, or focal chest signs on auscultation [Reference von Baum25]. Ethical approval for the investigations was obtained from the ethics committees of the local clinical centres. Respiratory tract specimens (sputum, bronchoalveolar lavage fluid) and serum samples were taken in parallel from patients with symptoms of pneumonia and processed in an authorized diagnostic centre to standardize the sample treatment [Reference Welte, Suttorp and Marre24]. Sampling was carried out between 1 and 4 days after onset of symptoms. All infections investigated occurred as single cases; none was part of a disease cluster or an outbreak of M. pneumoniae infections. The use of antibiotics before sampling was excluded. Frozen aliquots of the serum samples and of extracted DNA of the sputum samples were sent to the Institute of Medical Microbiology and Hygiene at Dresden University (German reference laboratory for Mycoplasma and Ureaplasma of the Robert Koch Institute) for specific diagnosis of M. pneumoniae infections.

Fig. 1. Occurrence of different subtypes and variants of M. pneumoniae in the six local centres in Germany investigated within the CAPNETZ study between 2003 and 2006. Data based on molecular typing of 18 (centre 1), 21 (centre 2), 13 (centre 3), 24 (centre 4), 13 (centre 5) and 45 (centre 6) M. pneumoniae-positive sputum and bronchoalveolar lavage fluid samples of adult pneumonia patients.
Detection of M. pneumoniae and subtyping
DNA of respiratory tract samples was pre-screened using the real-time PCR approach targeting a conserved inter-repetitive region of the P1 adhesin [Reference Dumke26]. Subsequently, M. pneumoniae-positive samples were subtyped independently of culture as described previously [Reference Dumke22]. Briefly, the nested-PCR procedure amplifies a part of the repetitive element RepMp2/3 within the P1 gene showing characteristic and conserved sequence differences between the known subtypes and variants of M. pneumoniae. Sequencing of the PCR product allows reliable classification of the strain colonizing the respiratory tract of the patient.
Serological examinations
Selected sera of patients with a known genotype of M. pneumoniae in the respiratory tract sample were tested for M. pneumoniae-specific IgA, IgG and IgM antibodies with a commercial ELISA procedure (Genzyme Virotech, Germany) according to the manufacturer's recommendations. By using internal control sera the calculated cut-off for the positive reaction of a serum was set to >11·0 test units. Sera with test units between 9·0 and 11·0 were considered as borderline. In addition, to increase the specificity of the serological approach all ELISA-positive sera were investigated with a commercial Western blot test (Genzyme Virotech).
To detect antibodies against the subtype- and variant 2a-specific regions of the P1 adhesin, recombinant proteins were used as antigens for an in-house ELISA as previously described [Reference Dumke, Schurwanz and Jacobs23]. Briefly, the recombinant proteins rpall (positive control, conserved near C-terminal region of the P1 protein), rpST1 (subtype 1-specific), rpST2/V2 (subtype 2- and variant V2a-specific), rpST1/ST2 (subtype 1- and subtype 2-specific) and rpV2 (variant V2a-specific) were expressed, purified and concentrated. Due to the location of the sequence differences between the subtypes and variants in the repetitive elements RepMp4 and RepMp2/3 of the different P1 genes, the subtypes and the variant 2a are in each case characterized by two recombinant proteins. The recombinant proteins were applied as antigens (5 μg/ml carbonate buffer) in a conventional ELISA test to detect subtype- and variant-specific antibodies in 1:100 diluted sera of pneumonia patients. Peroxidase-conjugated anti-human IgA and anti-human IgG (Sigma, USA; 1:500) were used as secondary antibodies. The available sera against the recombinant proteins rpST1, rpST2/V2, rpST1/ST2 and rpV2 [Reference Schurwanz, Jacobs and Dumke27] were used to prove the cross-reactivity of the sera with common bacterial agents of community-acquired pneumonia (Legionella pneumophila ATCC 33152, Moraxella catarrhalis ATCC 23246, Chlamydophila pneumoniae TW-183, Streptococcus pneumoniae ATCC 6305, Haemophilus influenzae ATCC 49247) and human HeLa cells in ELISA tests.
RESULTS
Molecular subtyping based on the amplification of repetitive element RepMp2/3 located in the P1 gene of M. pneumoniae resulted in 134 analysable sequences. No sequences typical of subtype 3 and variant 1 strains were detected from the CAPNETZ investigations within the time period 2003–2006 in Germany. In summary, data differences between the distribution of the subtypes and variants characterized in the six local centres were observed (Fig. 1). For example, variant 2a strains of M. pneumoniae dominated in the samples investigated in centre 2 (43%) whereas between 15% and 31% of variant 2a strains were found in the other centres. Variant 2b isolates were detected in five of the six centres with rates between 5% and 15%. In centres 1 and 4, subtype 1 strains dominated with proportions of 58% and 44%, respectively. Most of the sequences found in centres 3, 5 and 6 (36–46%) could be assigned to subtype 2.
The time-dependent distribution of the subtype- and variant-specific sequences of M. pneumoniae between 2003 and 2006 showed an increase of variant 2a and variant 2b strains (Fig. 2). In 2006, variant 2a isolates for the first time represented the most frequent type of M. pneumoniae (35%) whereas 9% of all strains belonged to the variant 2b genotype. A clear dominance of one of the two subtypes was not found in the investigation period. Summarizing the typing results from the years 2003 to 2006, an almost equal distribution of subtype 1 (31%) and subtype 2 bacteria (35%) was observed.
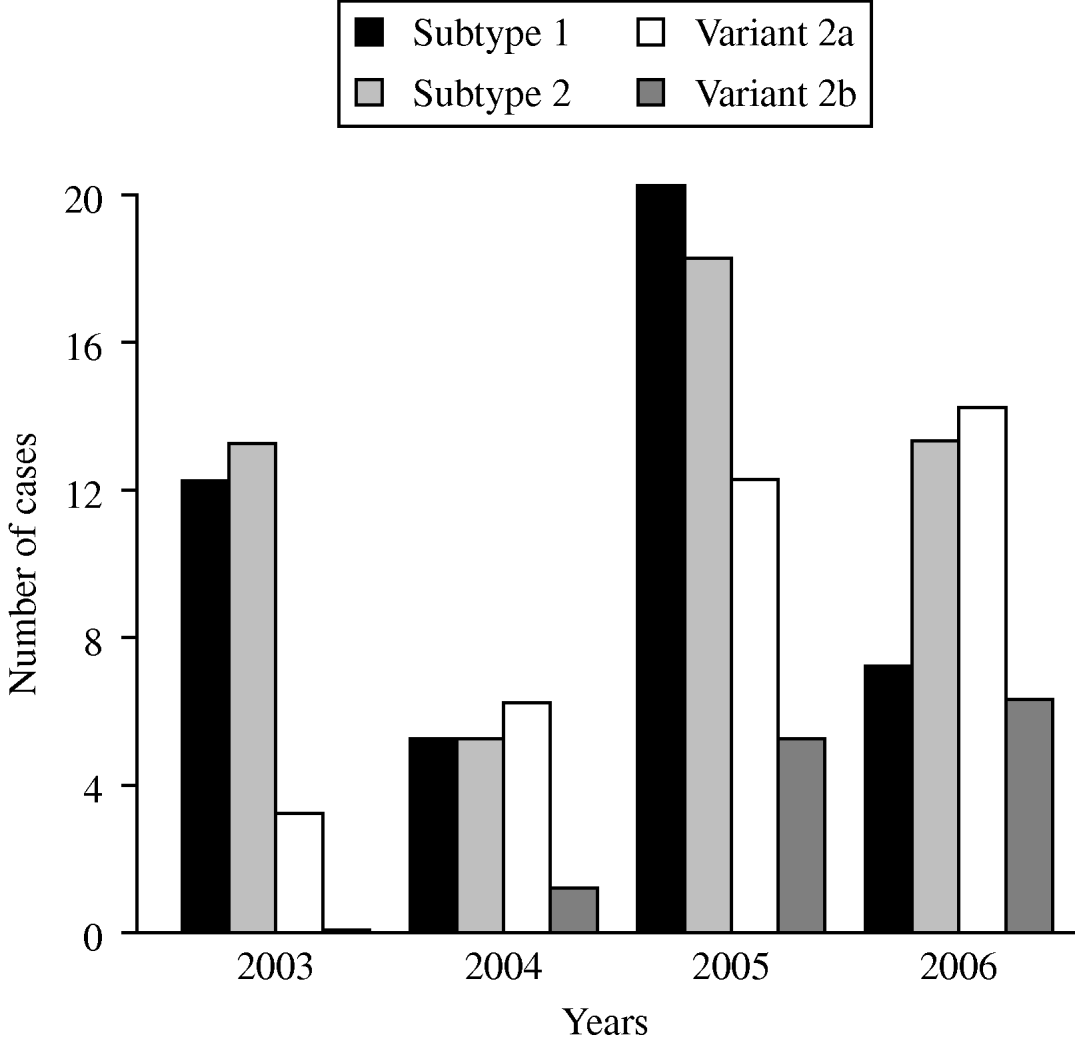
Fig. 2. Time-dependent distribution of subtype- and variant-specific sequences of M. pneumoniae in sputum and bronchoalveolar lavage fluid samples of patients with symptoms of community-acquired pneumonia within the CAPNETZ study.
Twenty-seven sera of pneumonia patients were tested to estimate the occurrence of antibodies against the variable regions in the P1 protein in pneumonia patients with a known genotype of M. pneumoniae in the respiratory tract specimen. Nine sera each were available with subtype 1, subtype 2 and variant 2a strains in the respiratory tract. Using the commercial ELISA test to detect antibodies to M. pneumoniae, all 27 sera were IgA-positive (arithmetic mean of the calculated test units ±standard deviation 33·0±26·8, min.–max. 11·1–118), 24/27 sera (89%) were IgG-positive (arithmetic mean of the calculated test units in positive samples 20·6±10·7, min.-max. 12·1–54·7) and 17/27 sera (63%) were IgM-positive (arithmetic mean of the calculated test units in positive samples 26·9±8·9, min.-max 14·8–42·9). The results of testing the 27 sera with the in-house ELISA using purified recombinant proteins derived from the conserved and subtype- and variant 2a-specific regions of the P1 protein as antigens are summarized in Figure 3. Because of the results using the commercial ELISA test, the investigations were limited to the detection of IgA and IgG antibodies. Highest mean optical density (OD) values regarding both of the investigated IgA (1·8±0·8) and IgG (2·0±1·2) antibodies were obtained for the conserved C-terminal part of the P1 adhesin representing the positive control. The results of ELISA reactivity of the four subtype- and variant 2a-specific recombinant proteins with sera of pneumonia patients with a confirmed genotype of M. pneumoniae revealed that despite the fact that certain reactions of genotype-specific antigens with type-specific sera corresponding to these regions showed higher OD values compared to the other antigens (e.g. IgA reaction of rpST1 with sera of patients with subtype 1-specific sequences in the respiratory tract) no statistically significant differences between the reaction of the type-specific recombinant proteins with the level of IgA and IgG antibodies in sera of the investigated patients could be measured (t test, P<0·05). The sera of ten pneumonia patients who tested negative with the commercial M. pneumoniae ELISA resulted in OD values <0·5 for all five recombinant proteins used in the study (data not shown). This value can be considered as the cut-off of the test procedure. Furthermore, no cross-reactivity of the four genotype-specific recombinant proteins used in the study with five bacteria species commonly causing community-aquired pneumonia and HeLa cells could be observed (data not shown).
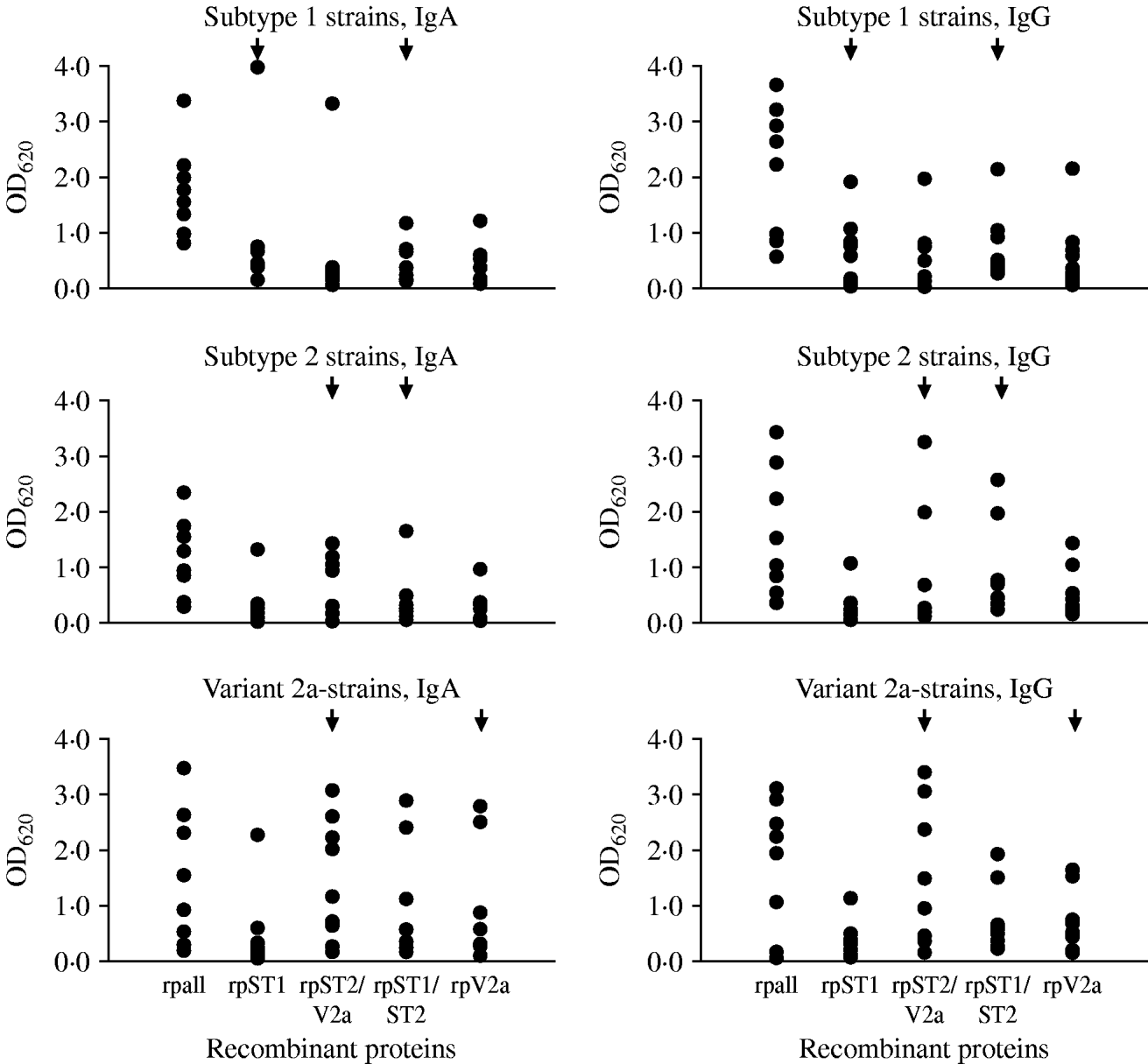
Fig. 3. Results of the in-house ELISA test using recombinant proteins to detect IgA and IgG antibodies to the conserved part (rpall) and to subtype- and variant-specific regions of the main P1 adhesin of M. pneumoniae in sera of pneumonia patients with genotyped M. pneumoniae strain in the respiratory tract. Arrows indicate the recombinant proteins which are derived from the specific regions of the given genotype of M. pneumoniae.
DISCUSSION
In a recent report, M. pneumoniae clinical isolates were investigated with a multi-locus variable number tandem repeat (VNTR) analysis and 26 distinct VNTR types were found [Reference Dégrange28]. Nevertheless, regarding the characterized proteins which are important for the host immune response, M. pneumoniae is a very homogeneous species showing remarkable sequence differences in only a few genes [Reference Dumke19]. Most of the variations could be found within the repetitive elements RepMp1, RepMp 2/3, RepMp4 and RepMp5 [Reference Ruland, Wenzel and Herrmann29] occurring in 8–14 copies over the entire length of the genome of the sequenced strain M. pneumoniae M129 [Reference Himmelreich30]. Since 1990, patient isolates of M. pneumoniae have been classified into subtypes 1 and 2 according to sequence differences in the two repetitive regions RepMp2/3 and RepMp4 in the gene coding for the main P1 adhesin [Reference Su31]. The appearance of new subtypes can be explained by recombination events within these repetitive elements. Nevertheless, the number of subtypes described up to now in patient isolates is much lower than can be expected theoretically from the different copies of repetitive elements in the genome. The first patient isolates demonstrating sequence differences in the repetitive element RepMp2/3 of the P1 gene were reported from Japan, whereas the RepMp4 copy was in accordance with the corresponding repetitive region in subtype 2 strains [Reference Kenri32]. These isolates were later denoted as variant 2a. The counterpart of the 2a strains was characterized in 2001 as showing sequence variations in RepMp2/3 in combination with a subtype 1-specific RepMp4 (variant 1 [Reference Dorigo-Zetsma33]). The description of these strains led to the classification that a subtype is defined by sequence differences in both of the repetitive elements in the P1 gene whereas a variant is characterized by varying regions in one of the repetitive regions only [Reference Dumke34]. Since culturing of M. pneumoniae is time-consuming and insensitive, subtyping has recently been carried out by amplification and molecular characterization of PCR products directly from respiratory tract samples [Reference Kenri18, Reference Dumke22]. The molecular approach allows the reliable culture-independent classification of all subtypes and variants of M. pneumoniae described so far and the rapid subtyping of strains in a high number of patients, and avoids an underestimation of genotypes due to possible differences in the propagation efficiency of subtypes and variants in culture.
Sequencing of the variable RepMp2/3 region of the P1 gene resulted in the detection of new variant 2b symptoms of community-acquired pneumonia in several patients in the USA [Reference Schwartz21], Germany and Switzerland [Reference Dumke22]. Nevertheless, the epidemiological evaluation of the worldwide distribution of the different subtypes and variants of M. pneumoniae is hampered by the limited number of subtyping studies. Most of the reports divided the isolated patient strains into the two subtypes only [Reference Pereyre20], allowing no interpretation of the epidemiological importance of the variants of M. pneumoniae described. Two studies included the known variants and found an increase in the occurrence of variant 2a isolates in Germany and Japan in recent years [Reference Kenri18, Reference Dumke22]. Variant 2b strains were detected in central Europe by molecular methods only and have not been cultured to date. A single isolate from Denmark was characterized as variant 1, indicating the occurrence of genotypes with a very limited distribution in the human population. The emergence and stable distribution of patient isolates of M. pneumoniae showing novel regions within the adhesins recombined from the reservoir of the different repetitive elements seems to be a rare event. In a recent study from France, the occurrence of an unknown subtype was described in one of 155 investigated patient strains collected between 1994 and 2006 [Reference Pereyre20]. According to the limited detection in the available typing studies, it is difficult to assess the epidemiological significance of both of these subtype and variant strains.
The current study displays differences in typing results between the local centres involved, indicating the problem of accurate detection of the dominant strains that cause disease symptoms. However, since all infections were sporadic cases a bias due to clusters of infections could be excluded. A country-wide survey should include the results of more than one or two regions in order to give an overall insight into the type-specific epidemiology of infections due to M. pneumoniae.
Despite the results of the subtyping studies performed, the epidemiological significance of the time-dependent circulation of different subtypes and variants of M. pneumoniae is not fully understood. Up to now no differences between the known subtypes and variants of M. pneumoniae in terms of severity of illness or the localization of the symptoms within the upper and lower part of the human respiratory tract have been reported. Moreover, a relationship between the detection of a particular genotype of M. pneumoniae and the age of the affected patients or the symptoms of the infection could not be found in the current study. It can be hypothesized that the described changes in the nucleotide sequence and the resulting differences in the proteins have limited influence on the pathogenicity of the subtypes and variants. The sequence differences in the main P1 adhesin are regarded as important for the interaction of M. pneumoniae with the host immune system since the protein was identified early on as the most immunogenic antigen of the bacterium [Reference Hu35]. Experimental data suggested an influence of the host immune system on the colonization efficiency with a distinct subtype or variant of M. pneumoniae [Reference Jacobs17, Reference Dumke34]. Furthermore, the subtype- and variant-specific regions of the P1 adhesin were recently characterized as antigenic [Reference Dumke, Schurwanz and Jacobs23]. These experimental results support the hypothesis that the described epidemiological pattern of an increase in infections due to M. pneumoniae within time intervals of 3–7 years [Reference Lind12, Reference Eun13] might be explained by time-dependent changes in the circulation of subtype-specific antibodies in the human population. The results of a long-term study demonstrated the changeover of periods with an increased type shift from subtype 1 and subtype 2 and vice versa followed by a period of strict dominance of one subtype in an almost 5-year interval in Japan [Reference Kenri18]. The switch from the dominance of subtype 1 to subtype 2 strains between 2000 and 2005 was associated with a significant increase in the Mycoplasma pneumonia cases in this period. In contrast, the present report showed the stable occurrence of both subtypes of M. pneumoniae in the 2003–2006 period without a pronounced dominance of a distinct genotype. This is in accord with the results of subtyping clinical isolates in France, which confirmed the almost equal distribution of types 1 and 2 between 1998 and 2006 [Reference Pereyre20]. Further investigations are necessary to clarify the time-dependent occurrence of M. pneumoniae genotypes in the human population and the influence of these changes on the incidence of diseases.
Regarding the role of the host immune response for the subtype switch, the CAPNETZ network gave the unique opportunity to investigate a number of sera of adult patients with a known subtype of M. pneumoniae in the respiratory tract. This offers the possibility of comparing the quantitative occurrence of subtype- and variant-specific antibodies with the genotype responsible for the actual infection. The detection of specific antibodies in patient sera is still a common assay used to confirm diseases due to M. pneumoniae in routine diagnostic laboratory procedures. The development of IgA antibodies after an infection with M. pneumoniae is typical for adult patients and can be considered as the consequence of a current infection, whereas about 20% of adults did not show a detectable IgM response [Reference Thacker and Talkington36]. However, the diagnostic value of elevated IgA antibody titres in the acute phase of M. pneumoniae infections is uncertain, since specific IgA antibodies can also be detected in healthy persons [Reference Csango, Pederson and Hess37] indicating specificity problems of the test procedures or the persistence of IgA antibodies after resolution of clinical symptoms. Regarding the serological data of the curent study, the epidemiological value of the IgG antibodies arises from past infections with a distinct subtype or variant of M. pneumoniae [Reference Hammerschlag38] and the resulting type-specific antibody response, which could influence the actual reinfection. As expected, the sera of M. pneumoniae-positive patients showed the highest OD values after reaction with the highly immunogenic C-terminal part of the P1 adhesin [Reference Svenstrup39, Reference Chaudhry40]. Using sera of animals infected intranasally with M. pneumoniae subtype 1, subtype 2 or variant 2a strains, the investigation of the four subtype- or variant 2a-specific recombinant proteins resulted in a type-specific immune response with significant differences between the antigens [Reference Dumke, Schurwanz and Jacobs23]. In contrast, the results of investigation of single serum samples of patients with confirmed M. pneumoniae pneumonia indicated that the occurrence of antibodies in the serum samples against the subtype- and variant-specific protein regions was not strongly correlated with the genotype of the strain in the respiratory tract of the patient. This means that neither the mean specific IgA values that reflect the immune response to the M. pneumoniae-type of the actual acute-phase infection nor the low type-specific IgG levels resulting from previous infections favour re-infection with a given genotype of M. pneumoniae. This fact might be explained to some extent by the time-dependent development of antibodies in the sera of infected patients [Reference Nilsson, Björkman and Persson41] and by the possible multiplicity of previous infections complicating the differentiation of subtype- and variant-specific IgG antibodies.
In conclusion, the results of the current study confirm the occurrence of different, but not all, known subtypes and variants of M. pneumoniae during a standardized 4-year study period in Germany. Whereas subtypes 1 and 2 strains occurred in an almost equal distribution, the rate of variant 2a and 2b isolates increased. Recording of the real distribution of the M. pneumoniae genotypes in a human population was complicated by marked regional differences in type pattern. The missing correlation of the mean subtype- and variant-specific IgA and IgG antibody concentrations in sera of pneumonia patients with a confirmed genotype in the respiratory tract did not support the hypothesis of an influence of host immune response on the colonization efficiency of a distinct M. pneumoniae subtype or variant in the respiratory tract in comparison to other genotypes.
ACKNOWLEDGEMENTS
This work was supported by the BMBF network of competence (CAPNETZ). We gratefully acknowledge the excellent laboratory work of Carolin Dix and Kerstin Riedel.
DECLARATION OF INTEREST
None.







