INTRODUCTION
Shiga toxin (Stx)-producing Escherichia coli (STEC) is a foodborne enteric pathogen that causes human illness ranging from mild diarrhoea to life-threatening haemolytic uraemic syndrome (HUS) [Reference Karmali, Gannon and Sargeant1]. Epidemiological studies have shown that STEC can potentially enter the human food chain from a number of animal sources; the most common sources are unpasteurized dairy products and meats cross-contaminated with faeces or intestinal contents after slaughter [Reference Hussein and Sakuma2].
Because most causes of STEC outbreaks in the USA have been traced to beef containing the STEC serotype O157:H7 [3], epidemiological studies have focused on the prevalence of this serotype in beef and beef cattle. However, additional STEC serotypes, including serotypes O26, O91, O103, O111, O118, O145, and O166, have been isolated from beef and have caused human illnesses worldwide [4]. In Japan, the dominant serotype associated with human STEC infection is O157:H7, followed by O26:H11 and O111:H−, with these three accounting for more than 95% of human STEC infections [5]. Of the 373 STEC serotypes isolated from cattle faeces or hides, 65 were identical to those detected in HUS patients, and 62 are known to cause other human illnesses [Reference Hussein6]; these data highlight the fact that cattle and bovine food products are important reservoirs for human STEC infection.
Notably, in some Asian countries, it is customary to eat bovine offal products including liver, heart, and intestines; some are consumed raw, as so-called sashimi. Recent epidemiological data have indicated that of the 28 foodborne STEC O157 outbreaks in which their causative foods were identified in Japan during 2007–2010, 11 (39%) outbreaks were caused by the consumption of these bovine offal products [7]. These cases suggest that such food products might be one of the major sources of foodborne STEC infection in Japan. Despite this understanding, contamination rates and doses of STEC in those food products along their supply chain remain unclear, although a small-scale detection study has been reported [Reference Kanki8].
In the present study, we examined the prevalence of STEC (particularly serotypes O157, O26, and O111) in various types of bovine offal products that are widely distributed in Japan. Throughout the comparative assessment of the growth kinetics of STEC, we found that these food products significantly affected the growth of STEC during enrichment. Therefore, we investigated background microbiota from the offal products and their effects on the growth of STEC.
MATERIALS AND METHODS
Food samples
A total of 229 raw bovine offal products that are commercially distributed throughout Japan via the internet were purchased from July to November 2010 (Table 1). These samples were delivered to the laboratory by rapid, cold road transport and were processed on arrival. These samples included liver (n=36), rumen (n=21), reticulum (n=22), omasum (n=38), abomasum (n=24), small intestine (n=54), large intestine (n=22), heart (n=6), and others (two kidney and four mixed intestinal products).
Table 1. Prevalence of stx and O157, O26, O111 serotype-specific genes in bovine internal organ foods

* Others represent kidney (n=2) and mixed intestinal products (n=4).
Culture enrichment and DNA extraction
A 25-g portion of each raw sample was diluted with 225 ml buffered peptone water (BPW) (pH 7·4) (Oxoid, UK) in a Stomacher bag (Eiken Kagaku, Japan), followed by homogenization for 1 min at 5·0 strokes/s using a Lab blender (IUL Instruments, Spain). After incubation at 42°C for 20 h, a 1 ml aliquot of the enrichment broth was centrifuged at 21 500 g for 5 min, and the cell pellets were resuspended in 100 μl PrepMan Ultra solution (Life Science Technology Inc., USA) to extract DNA according to the manufacturer's instructions.
PCR detection for stx and serotype-specific genes
The DNA extract prepared as described above was subjected to PCR to detect the Shiga toxin (stx) gene as described previously [Reference Lin9]. The stx-positive samples were further subjected to PCR to detect the following genes: rfbE O157 [Reference Desmarchelier10], wzy O26 [Reference DebRoy11], and rfb O111 [Reference Paton and Paton12], which can specifically detect serotypes O157, O26, and O111, respectively.
Isolation and characterization of STEC from bovine offal products
Bacterial cultures that were PCR positive for any of the three serotype-specific genes were subjected to immunomagnetic separation using Dynabeads specific for O157 or O26 (Dynal, Norway) (according to the manufacturer's instructions) to isolate the target serotype bacteria. The following selective agar media were used: KBM STEC Chrom agar (Kojin Bio, Japan) and cefixime tellurite-sorbitol MacConkey (CT-SMAC) agar (Eiken Kagaku) for O157 and KBM STEC Chrom agar (Kojin Bio) and cefixime tellurite-rhamnose MacConkey (CT-RMAC) agar (Eiken Kagaku) for O26. The STEC isolates that were finally obtained were serotyped and genetically characterized for stx, eaeA, EhlyA and uidA as previously described [Reference Feng and Monday13]. Stx production was assayed with VTEC-RPLA (Denka Seiken, Japan).
Construction of kanamycin (Km)-resistant STEC O157
The plasmid pWM1007, which contains kan (Km-resistant) loci [Reference Miller14], was introduced into STEC O157 strains 204, 466, and 470 by electroporation. Successful transformants (designated 204-Km, 466-Km, 470-Km, respectively) (Table 2) were selected on CT-SMAC agar containing 50 μg/ml Km (CT-SMAC-Km).
Table 2. Bacterial strains used in the study
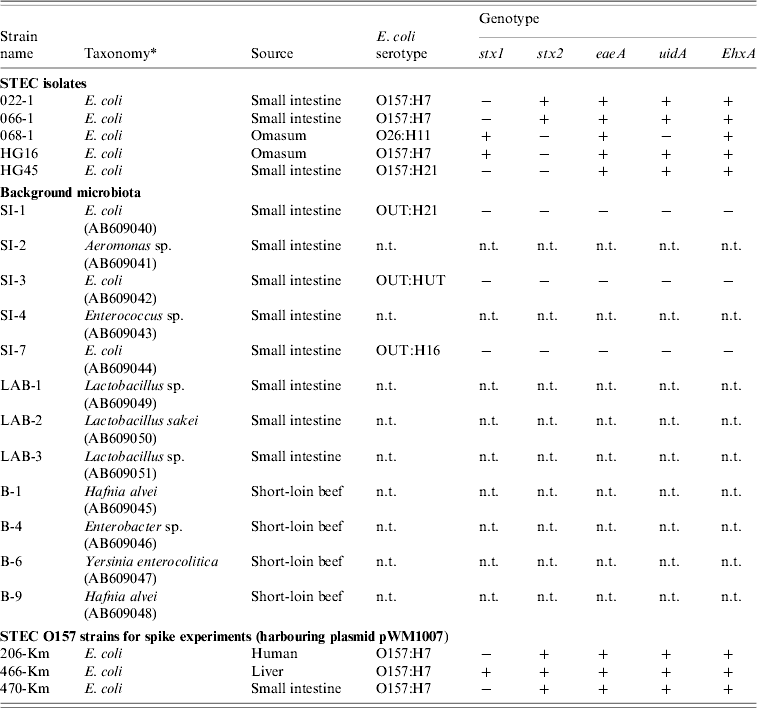
n.t., Not tested.
All E. coli were genotyped for the presence (+/−) of stx, eaeA, uidA, and EhxA genes, and were O- and H-serotyped.
* Taxonomy of the background microbiota were based on the 16S rRNA sequences (GenBank accession numbers given in parentheses). Km-resistant STEC O157 strains 204-Km, 466-Km, 470-Km were constructed by introducing the plasmid pWM1007.
Viable cell count of STEC O157 in bovine food homogenates
The STEC O157 strain 204-Km was grown in LB medium supplemented with 50 μg/ml Km (LB-Km) at 37°C for 20 h. After being washed with sterile PBS twice, 100 μl each of the serially diluted suspension were separately used to inoculate 5-g portions of short-loin beef, liver, and small-intestine products in a Stomacher bag (Eiken Kagaku). Following suspension in 45 ml BPW, homogenization was performed in a Lab blender (IUL Instruments) at 5·0 strokes/s for 1 min. The homogenates were then incubated at 42°C for 20 h. The numbers of viable O157 cells were determined by plating the homogenates onto CT-SMAC-Km at 0 h and 20 h post-incubation. To exclude strain-specific growth characteristics, we separately inoculated three O157 strains (204-Km, 466-Km, 470-Km) in bovine small-intestine products and their growth through BPW enrichment was also measured by plate count.
Measurement, isolation, and characterization of background microbiota in bovine offal
A 5-g portion of small-intestine product and short-loin beef was homogenized (1 min, 5·0 strokes/s) in 45 ml BPW as described above, and the serial dilutions were immediately plated on Nutrient agar (NA) plates (Eiken Kagaku) to determine aerobic plate counts (APCs) by incubation at 37°C for 22 h, or MRS agar (BD Biosciences, USA) to measure the number of lactic acid bacteria (LAB) by anaerobic incubation using GasPak (BD Biosciences) at 37°C for 48 h as described previously [Reference Vold15]. The remainder of the homogenates were thereafter incubated at 42°C for 20 h, and also subjected to the same measurement of bacterial counts. To rule out bacteria that predominantly grew in the homogenates after incubation, six representative colonies grown on the NA and MRS plates from the 20-h culture were separately chosen, and their 16S rRNA sequences were determined by cycle sequencing using the primers 5′-CAGGCCTAACACATGCAAGTC-3′ and 5′-GGGGGGTGTACAAGGC-3′, in an ABI3730xl DNA analyser (Life Science Technology) according to the manufacturer's instructions. In the case of complete sequence similarity in the isolates from NA plates, one representative isolate was biochemically characterized with the Api-20E kit (bioMérieux, France) and used in the following studies.
Competitive growth assay
Co-incubation with background microbiota complex
A 5-g portion of short-loin beef or small-intestine samples was suspended in 45 ml BPW and vortexed for 60 s. Subsequently, the suspensions were filtered through a 20-μm-diameter filter (Millipore, USA) to remove the majority of the microbes from the food matrices. The filtrates were then centrifuged for 15 min at 2150 g, and the bacterial pellets were carefully washed and resuspended in 45 ml fresh BPW. About 1·5×103 cells of STEC O157 strain 204-Km were then used to inoculate background microbiota-containing BPW suspensions in a Stomacher bag and these cultures were then incubated at 42°C for 20 h. After incubation, the growth of STEC O157 was determined by plate counts on CT-SMAC-Km.
Co-incubation with representative microbiota in food homogenates
A 5-g portion of short-loin beef or small-intestine products was added to 45 ml BPW, homogenized as described above in a Stomacher bag, followed by incubation for 30 min at 63°C (heat+) to kill most bacteria, or kept at room temperature (heat–) not to kill bacteria (we confirmed that following the above heating, both types of food homogenates contained <10 c.f.u./ml bacteria, enumerated by colony counts on NA agar). After cooling down to room temperature, the food homogenates were inoculated with 8·6×102 cells of O157 strain 204-Km and co-cultured with approximately 102 c.f.u. orders of representative background microbiota from small-intestine products (SI-1, -2, -3, -4, and LAB-2) at 42°C for 20 h. The growth of O157 cells was determined by plate count on CT-SMAC-Km. The food homogenates spiked only with O157 culture (without microbiota) were used as controls.
Nucleotide sequence data
The 16S rRNA sequences from the representative background microbiota were deposited in GenBank under accession numbers AB609039–AB609051.
Statistical analysis
Data for the plate counts represent means±standard deviation from three sets in two independent experiments unless otherwise indicated. Statistical significance was calculated by Student's t test (P<0·05).
RESULTS
Prevalence of STEC-related genes in bovine offal products in Japan
From July to November 2010, 229 bovine offal products that were commercially distributed throughout Japan were subjected to testing for the prevalence of STEC. PCR screening detected the stx gene in 38 (16·6%) samples. These samples included heart (33·3%), liver (11·1%), rumen (23·8%), reticulum (27·3%), omasum (10·5%), abomasum (8·3%), small intestine (18·5%), large intestine (18·2%), and others (0%) (Table 1). Of these samples, eight were positive for rfbE O157, and three were positive for wzy O26. One omasum sample was positive for both rfbE O157 and wzy O26 (Table 1). The rfb O111 gene was not detected in any of the samples tested.
Isolation and characterization of STEC O157 and O26
Of the 10 samples that were positive for rfbE O157 or/and wzy O26, immunomagnetic separation in combination with the selective media enabled us to isolate three O157:H7, one O157:H21, and one O26:H11 STEC from small-intestine and omasum products (Table 2). All STEC strains except the strain HG45 (serotype O157:H21) produced cytotoxic Shiga toxins, and their stx genotypes consisted of stx1 alone (n=2), and stx2 alone (n=2) (Table 2). They all possessed the EhxA gene, and the uidA gene was detected in all O157 isolates (Table 2).
Type of food matrix that affects the growth kinetics of STEC O157
Having little information on the enrichment efficiency of the bovine offal products for STEC bacteria, we experimentally used the Km-resistant STEC O157 strain 204-Km to inoculate samples of short-loin beef, liver, and small intestine, followed by homogenization. After incubation of the homogenates, the growth of O157 cells was determined. The short-loin beef homogenates allowed the STEC bacteria to grow to a concentration ranging between 1·1×107 and 2·8×107 c.f.u./ml when inoculated with 2·5×103 c.f.u. of STEC O157 strains (Fig. 1 a). However, the inoculums were not significantly increased in the homogenates of small-intestine products, exhibiting only a 3·2- to 4·8-fold increase (8·1×103 to 1·2×104 c.f.u./g) (Fig. 1 a). The growth of O157 cells in the liver sample homogenates was intermediate; 5·9×104 to 3·2×105 c.f.u./g of STEC O157 were recovered under the same conditions (Fig. 1 a). The reduced extent of STEC O157's growth in the homogenates of bovine small-intestine products was confirmed not to be a strain-dependent characteristic because three different O157 strains exhibited similar trends towards O157 growth (Fig. 1 b). Taken together, the data demonstrated that homogenates of bovine digestive-tract products significantly reduced the extent of STEC O157 growth in the enrichment process compared to homogenates of loin beef and bovine liver products.
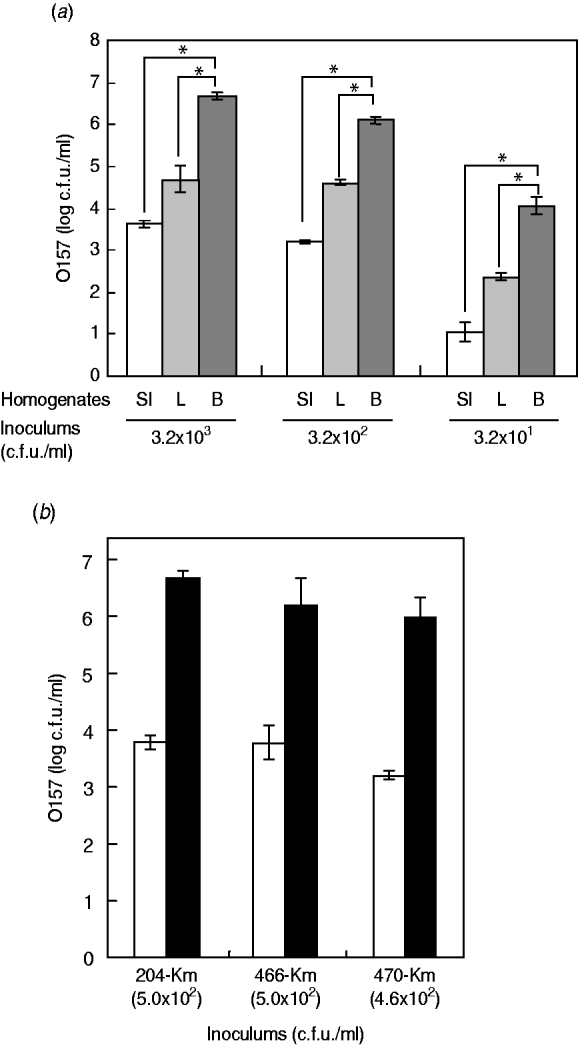
Fig. 1. Food matrixes of bovine digestive-tract products significantly affect the growth of STEC O157 during the enrichment process. (a) Different numbers of STEC O157 strain 204-Km (3·2×103, 3·2×102, 3·2×101 c.f.u.) were inoculated into 5-g portions of bovine products (SI, small intestine; L, liver; B, short-loin beef) and homogenized in BPW. After incubation, the STEC O157 cells were counted on CT-SMAC-Km. (b) Three STEC O157 strains (204-Km, 466-Km, 470-Km) were used to inoculate bovine small-intestine (□) and short-loin beef (▪) products and homogenized in BPW. In panels (a) and (b), means represent viable numbers of O157 (c.f.u./ml), and error bars represent standard deviation. Asterisks indicate statistical significance (P<0·05).
Background microbiota in bovine intestinal products plays a role in affecting STEC O157 growth in BPW
Due to the significant effect of the homogenates of small-intestine products on STEC O157 growth, we assumed that background microbiota in the homogenates might play a role in reducing the extent of growth of STEC bacteria in BPW. To examine this issue, we first isolated the background microbiota complexes from short-loin beef or bovine small-intestine products, using filtration-based isolation of the background microbiota; this enabled us to collect approximately 4·8×103 or 5·8×103 cells of the microbiota from 5-g short-loin beef and small-intestine products, respectively, which corresponded to 40% and 44·6%, respectively, of the APCs that were counted by direct plating of the homogenates (1·2×104 or 1·3×104 cells, respectively) (Fig. 2 a). Through the co-incubation with loin beef-originated background microbiota complex in the absence of food homogenates, the O157 strain 204-Km grew to a concentration ranging from 2·8×107 to 4·5×107 c.f.u./ml BPW medium (Fig. 2 b). In comparison, co-incubation with background microbiota from small-intestine products allowed STEC cells to grow only to 7·7×105 c.f.u./ml, which was about 30-fold less than from co-incubation with short-loin beef-derived background microbiota (Fig. 2 b). These data thus suggested that the background microbiota complex in small-intestine products played a role in affecting the growth of STEC O157 in BPW.
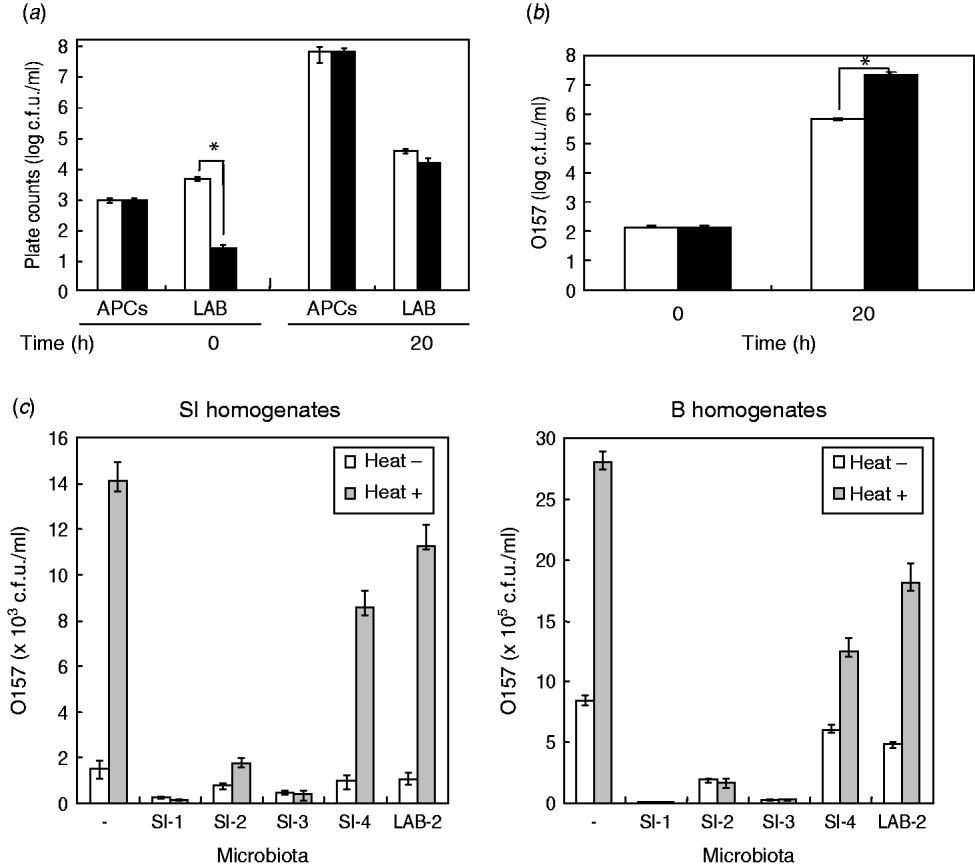
Fig. 2. Background microbiota affects the growth of STEC O157 during enrichment. (a) Aerobic plate counts (APCs) and lactic acid bacteria (LAB) counts in homogenates of small-intestine (□) and short-loin beef (▪) products were counted before (0 h) and after (20 h) incubation. (b) Background microbiota complex was isolated from small-intestine (□) or short-loin beef products (▪), then co-incubated with O157 strain 204-Km in BPW in the absence of food homogenates. At 0 h and 20 h post-incubation, numbers of STEC O157 were counted. Asterisks in section panels (a) and (b) indicate statistical significance (P<0·05). (c) Homogenates of small-intestine products (left panel) or short-loin beef (right panel) were treated with heat (heat+) for 30 min or untreated (heat–). These food sample homogenates were then inoculated with O157 strain 204-Km and representative microbiota from small-intestine products (SI-1, -2, -3, -4, and LAB-2). After incubation, the growth of O157 was determined. Food sample homogenates spiked only with O157 (with no spiked microbiota) were used as control (–). Error bars in panels (a)–(c) indicate standard deviation.
Bovine intestinal products contain different composition of background microbiota than short-loin beef products
Given the different effects of background microbiota from the loin-beef and bovine small-intestine products on the growth of O157 under the experimental co-culture conditions, we next examined the population and composition of APCs, and LAB in the homogenates of bovine short-loin beef and small-intestine products before and after incubation. Between the two types of the homogenates, APCs were not significantly different throughout the enrichment (1·2×103 and 1·3×103 c.f.u./ml at pre-incubation, and 7·8×107 and 7·1×107 c.f.u./ml at 20 h post-incubation, respectively) (Fig. 2 b). Moreover, the homogenates of small-intestine products contained more LAB compared to the short-loin beef sample homogenates at pre-incubation (6·8×103 and 4·2×101 c.f.u./ml, P<0·05), whereas both were increased to almost equal levels after incubation (Fig. 2 b). Through genetic and biochemical characterization, we subsequently identified representative bacterial colonies predominantly grown after incubation; the microbiota in small-intestine products consisted of E. coli, Aeromonas spp., and Enterococcus faecium (Table 2). In comparison, no E. coli were isolated from the short-loin beef homogenate sample, which instead contained Hafnia alvei, Enterobacter spp., and Yersinia enterocolitica (Table 2). In addition, representative LAB isolates from the homogenates of small-intestine products were genetically identified as Lactobacillus spp. and L. sakei (Table 2). These findings indicated that microbiota that grew predominantly in homogenates of bovine small-intestine products were compositionally different from those in the homogenates of short-loin beef.
Generic E. coli reduces the extent of STEC O157 growth in BPW homogenates
Having established that the background microbiota complex from bovine small-intestine products significantly reduced the extent of growth of O157 in BPW in the absence of food sample homogenates, and in which different composition of microbiota were predominantly grown compared to the homogenates of short-loin beef, we next examined the effect of representative microbiota isolates on STEC O157 growth by co-incubation with the food homogenates. Prior to incubation, the homogenates of small-intestine and short-loin beef products were treated with or without heating (63°C, 30 min). These homogenates were then spiked with STEC O157 and equal orders of microbiota isolates from small-intestine products, following by incubation for 20 h. In all homogenates, the heat treatment increased the growth of O157 compared to unheated homogenates (Fig. 2 c); in the absence of spiked microbiota, the heat treatment increased the number of O157 by 9·2-fold in small-intestine homogenates and by 3·3-fold in loin-beef homogenates, respectively (Fig. 2 c). Of homogenates spiked with microbiota, those with generic E. coli microbiota (SI-1, SI-3) allowed less growth of O157 cells in small-intestine and loin-beef homogenates (Fig. 2 c). Spiking with LAB (LAB-2) did not produce any marked influence on the growth of O157 in either type of homogenate, compared to generic E. coli (Fig. 2 c). Taken together, we were able to demonstrate that generic E. coli was one of the predominant microbiota in reducing the extent of STEC O157 growth in the homogenates during the enrichment process.
DISCUSSION
The types of food associated with STEC outbreaks and the geographical distribution of cases differ between countries. These differences have originated from local food preferences, culinary customs, and patterns of food distribution. Recent epidemiological data in Japan have revealed that consuming bovine offal products seems to be linked to the occurrence of STEC infection [7]. Therefore, for the first time, this study examined the prevalence of STEC in various types of bovine offal products.
The efficacy of a detection method depends on the choice of the base medium, selective agents, and their concentrations. The interactions between these factors are also expected to affect the sensitivity of the detection method, especially when the test sample contains a small number of STEC cells. After slaughter, bovine offal products are often treated with chlorine, and it is likely that such sanitizing damages or kills many of the STEC bacteria [Reference Farrell16]. Thus, it is conceivable that a portion of STEC in the test samples might have been damaged by chlorination. A comparative evaluation study on the recovery medium for STEC O157, O26, and O111 showed that BPW could efficiently recover damaged O157 bacteria culture, while tryptone soya and EC broths, and supplementation with vancomycin and novobiocin prevented bacterial growth [Reference Jasson17, Reference Drysdale18]. Further, BPW without additives was superior to BPW with additives for the isolation of STEC O157 by immunomagnetic separation from bovine faecal samples [Reference Foster19]. Therefore, we selected BPW without supplements as the enrichment media.
Many studies have focused the prevalence of STEC O157 in cattle faeces, hides, trim, or ground beef because of their epidemiological importance to public health [Reference Hussein6]. In this study, the stx gene was detected from a variety of bovine offal products. However, it is difficult to conclude that these rates reflect the contamination rates of STEC in these products because of the different numbers tested. Conversely, successful isolation of STEC O157 and/or O26 only from digestive-tract products including small-intestine and omasum (Table 2) suggested that these products might be contaminated with high populations of STEC pathogens, compared to the others. Most O157 STEC strains relating to human infection possess H7 flagellin or no flagellin (H−), while the serotype O157:H21 isolated from bovine small-intestine products in this study appears to be rare and the association with human infection is unknown, because no report exists for the isolation of this serotype from beef cattle and their products [Reference Hussein6].
We repeatedly subcultured the samples that were positive for stx and either rfbE O157 or wzy O26, in different enrichment media for the isolation of STEC. Nevertheless, some of the STEC were not isolated. Possible explanations for this result include the following: the STEC had already died, the STEC were minimally damaged during storage and transport of these products, or the growth of STEC was competitively inhibited by certain microbes present in the food samples during the processes of storage and enrichment.
Our focus on the growth kinetics of STEC in the food matrix during the process of enrichment thereby revealed that bovine small-intestine homogenates greatly reduced the extent of STEC O157 growth compared to short-loin beef homogenates. Consequently, we are able to demonstrate that in a variety of background microbiota, generic E. coli predominantly played a role in reducing the extent of STEC growth in the homogenates of bovine offal products. Although we also found higher levels of LAB in small-intestine products compared to that in short-loin beef, co-incubation with LAB weakly affected STEC O157 growth relative to generic E. coli and others in the food sample homogenates (Fig. 2 c). In addition, the numbers of LAB after incubation were not significantly different between the two types of food products and both reached only around 104 c.f.u./ml (Fig. 2 b), further supporting our notion that LAB might not be the predominant microbiota to affect the growth of STEC in the enrichment process. Even with heat pretreatment to kill most microbiota, O157 still grew more in homogenates of short-loin beef than in homogenates of bovine intestinal products (Fig. 2 c). This suggests the additional factors in the food matrices also influence the growth of O157. Although the bovine intestinal products contained greater amounts of fat content than the loin beef products (data not shown), a recent work by Bosilevac et al. reported that increased fat content did not interfere with recovery of STEC O157 from beef carcasses [Reference Bosilevac20]. Further exploration for such factors would be valuable to improve the isolation method of STEC from bovine offal products.
The population of STEC may also be affected by the processing and storage conditions of the food products. A previous study reported that feedlot cattle are infected with O157 at a level of about log 1·17 MPN/g, and some cattle exhibit a ~1000-fold increase at that stage of slaughter (log 5·66 MPN/g) [Reference Fegan21]. The samples tested in this study were further multi-processed with washing, cutting, packaging, storage and transportation at 4°C before being subjected to examination. The data provided here are only for the final products, and therefore the compositional changes of STEC and microbiota in these foods during each process needs to be clarified in our future work. A global metabolomic approach will be valuable to gain comprehensive information on the composition and functional roles of background microbiota in food products.
In conclusion, we demonstrated the prevalence of STEC in various types of bovine offal products for the first time; the prevalence data (i.e. contamination rates with the isolate's virulence characteristics) herein may be useful for the risk assessment of those products as a potential source of human infection. Because a low dose of this pathogen can cause a relatively high attack rate, even survival without growth in bovine digestive-tract products may pose a public health risk if the food is insufficiently heat-treated or consumed raw. Other data such as ‘annual consumption volumes/rates’ for these foods, would further improve our understanding of the epidemiological importance. The characteristic growth kinetics of STEC O157 shown in this study also indicates the importance of being aware when to test these foods.
ACKNOWLEDGEMENTS
We thank Shin-ichiro Yoshida (Japan Food Research Laboratories) and Taku Yamaguchi (Japan Frozen Food Inspection Cooperation) for technical support in bacterial detection. This work was financially supported in part by a Grant-in-Aid from the Food Safety Commission, Japan (No. 0805) and a Grant from the Ministry of Health, Labor and Welfare, Japan.
DECLARATION OF INTEREST
None.








