Introduction
Staphylococcus aureus is an important zoonotic pathogen with worldwide distribution. Due to the broad spectrum of virulence factors and the ability to develop antibiotic resistance, the infection of S. aureus results in a high morbidity and mortality [Reference David and Daum1–Reference Fu4]. In humans, S. aureus is usually associated with skin and soft-tissue infections, endovascular infections, pneumonia, septic arthritis, endocarditis, osteomyelitis and sepsis [Reference David and Daum1]. The colonisation of S. aureus in animals has been receiving comprehensive attention since animals may potentially act as a reservoir of human infection [Reference Aires-de-Sousa5, Reference Angen6].
Rabbit is one of the most important hosts of S. aureus. The infected rabbits are mainly characterised by subcutaneous abscesses, mastitis and pododermatitis [Reference Hermans7]. To our knowledge, S. aureus is widespread in rabbits in Fujian Province of China. However, knowledge about the epidemiology and characteristics of S. aureus in rabbit in Fujian Province is limited. In this study, multi-locus sequencing typing (MLST), tested virulence factors and antimicrobial susceptibility was done on different isolates of S. aureus recovered from lesions (pneumonia, mastitis and pododermatitis) in dead rabbits.
Methods
Sample collection and S. aureus isolation
From August 2017 to December 2018, 466 lung samples, 93 mastitis samples and 132 pododermatitis samples were collected from dead rabbits of nine rabbit farms in three cities (Fuzhou, Longyan and Nanping) of Fujian Province. Each sample was mixed with sterile phosphate buffer saline and homogenised to make 50% suspension. A 100 µl of suspension was inoculated on sheep blood agar plate for cultivation at 37 °C for 24 h. The suspected S. aureus colony was initially identified by colony morphology and Gram-staining. The presumptive colony was then sub-cultured and further confirmed by sequencing of 16S rRNA and nuc [Reference Weisburg8, Reference Brakstad, Aasbakk and Maeland9].
Multi-locus sequencing typing
Genotyping of S. aureus isolate was performed by MLST [Reference Enright10]. Briefly, seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi and yqiL) of S. aureus isolate were amplified and sequenced, and sequence type (ST) of the isolate was defined according to the allelic numbers of the seven housekeeping genes (http://www.mlst.net).
Virulence gene detection
The presence of virulence genes in S. aureus isolate was screened by PCR assays using primers previously reported, including thermonuclease (nuc) [Reference Brakstad, Aasbakk and Maeland9], panton-valentine leucocidin (PVL) toxin (pvl) [Reference Jarraud11], entorotoxin (sea and seb) [Reference Srinivasan12], toxic shock syndrome toxin-1 (tsst) [Reference Xie13], exfoliative (eta and etb) [Reference Xie13], haemolysin (hla and hlb) [Reference Jarraud11], clumping factor (clfA and clfB) [Reference Wang14] and fibronectin-binding protein (fnbpA and fnbpB) [Reference Wang14]. The PCR products were separated and sequenced.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of S. aureus isolates for 12 antibiotics was performed using the disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines [15]. The following antibiotics were used: penicillin, streptomycin, gentamycin, enrofloxacin, kanamycin, ciprofloxacin, cefminox, azithromycin, florfenicol, levofloxacin, ceftizoxime and ceftriaxone. The S. aureus ATCC 29213 was used as control. Moreover, the presence of mecA or mecC gene in S. aureus isolate was screened to confirm methicillin-resistant S. aureus (MRSA) [Reference Murakami16, Reference Paterson17].
Results
S. aureus isolation and identification
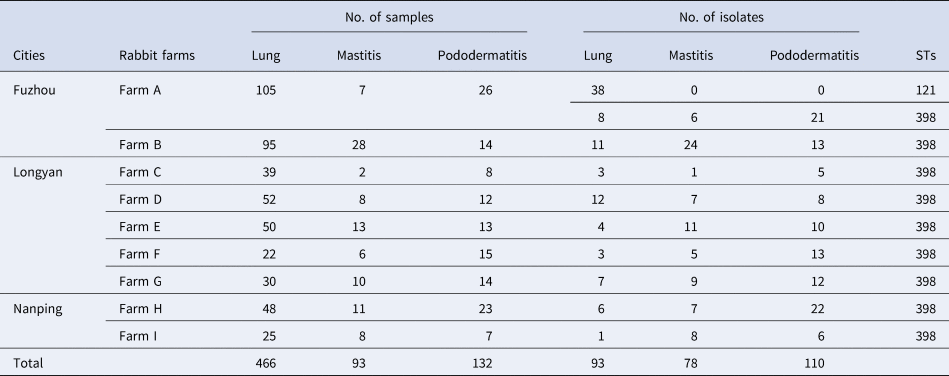
A total of 281 S. aureus isolates were recovered from the 691 samples of dead rabbits. Among them, 93 strains were isolated from 466 lung samples, 78 strains from 93 mastitis samples, 110 strains from 132 pododermatitis samples (Table 1).
Table 1. Sample collection, S. aureus isolation and sequence types of the isolates
Multi-locus sequencing typing
The genotype of S. aureus isolate was determined by MLST. It was shown that the 281 S. aureus isolates were divided into two STs, ST121 and ST398. In the genotype of ST121, all of the 38 isolates were only isolated from lung samples collected from Farm A (Table 1). In the genotype of ST398, 55 isolates were recovered from lung samples, 78 isolates from mastitis samples and 110 isolates from pododermatitis samples (Table 1).
All isolates were further clustered into clonal complexes (CCs) by using eBURST software (http://saureus.mlst.net/eBURST/). It was shown that the isolates were clustered into 2 CCs, CC121 and CC398 (Fig. 1).
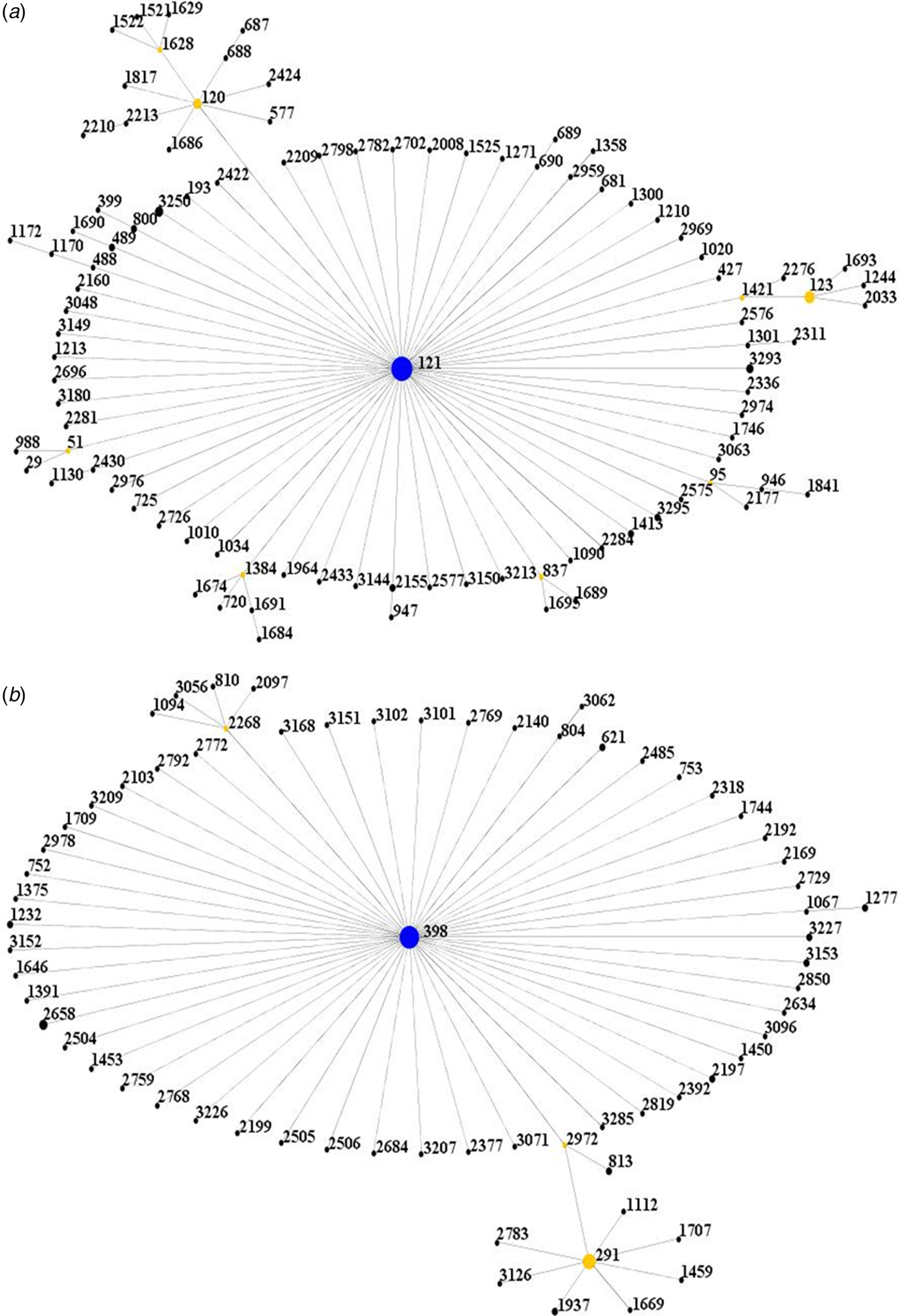
Fig. 1. Population snapshot of the isolates. (A) The ST121 is the putative founder of the CC121 and coloured blue. (1B) The ST398 is the putative founder of the CC398 and coloured blue.
Virulence gene detection
All the isolates were positive for the genes nuc, hla, hlb, clfA, clfB and fnbpA, whereas sea, seb, tsst, eta and etb genes were negatives. The pvl gene was detected in the 38 isolates of ST121 isolated from lung samples, and the fnbpB gene was positive in 109 isolates of ST398 recovered from lung, mastitis and pododermatitis samples.
Antimicrobial susceptibility testing
The antimicrobial susceptibility testing showed that all the isolates were susceptible to florfenicol, ceftizoxime and ceftriaxone. The resistance rates to penicillin, streptomycin, kanamycin and azithromycin were more than 10%. Sixteen strains were resistant to ⩾3 antibiotics and the mostly resistant antibiotics were streptomycin, kanamycin and azithromycin (Table 2). Notably, the ST121 isolates showed a lower number of resistances compared with ST398 isolates. Moreover, all isolates were mecA and mecC negative, indicating that these isolates were methicillin-susceptible S. aureus (MSSA).
Table 2. Antimicrobial susceptibility profiles of the isolates according to STs

P, penicillin; S, streptomycin; GM, gentamycin; ENR, enrofloxacin; K, kanamycin; CIP, ciprofloxacin; CFM, cefminox; AZM, azithromycin; FFC, florfenicol; LEV, levofloxacin; ZOX, ceftizoxime; CRO, ceftriaxone.
Discussion
This is the first study of the characteristics of S. aureus from rabbits in Fujian Province in southeastern China. The results showed that S. aureus was prevalent in the nine rabbit farms of Fuzhou, Longyan and Nanping. The mortality rates of the nine rabbit farms caused by the infection of S. aureus ranged from 18.37% to 52.9%, suggesting that S. aureus was the important pathogen causing the death of rabbits in these rabbit farms.
Generally, S. aureus has a combination of virulence factors, which were thought to contribute to the pathogenicity [Reference Edwards18]. All isolates in this study carried a panel of virulence genes nuc, hla, hlb, clfA, clfB and fnbpA. Moreover, the pvl gene was detected in the 38 ST121 isolates, and the fnbpB gene was detected in the 109 ST398 isolates. The 38 ST121 isolates that carried the pvl gene were all recovered from the lungs of dead rabbits with severe respiratory disease. The severe respiratory disease caused by the infection of the ST121 isolates was an isolated case on a rabbit farm in Fuzhou in late August 2017. The infection of the isolate caused the death of about 1000 rabbits in a 4-week period [Reference Wang14]. It was revealed that the PVL was related to necrotising pneumonia [Reference Lina19, Reference Labandeira-Rey20]. Infection of both PVL-positive MRSA and MSSA could lead to necrotising pneumonia [Reference Sicot21], indicating that the PVL of the 38 ST121 isolates might be one of the crucial factors contributing to the severe respiratory diseases. The fibronectin-binding proteins (FnBPA and FnBPB) are multifunctional virulence factors, which facilitate S. aureus colonisation and invasion of the host cells [Reference Edwards18, Reference Murai22]. In consistent with the previous reports, all ST398 isolates in this study expressed at least one fibronectin-binding protein, while some of the isolates expressed both FnBPA and FnBPB. A previous study showed that five out of the six ST398 strains isolated from raw milk of dairy cows with mastitis were positive of both fnbpA and fnbpB, while the other one isolate only carried fnbpA [Reference Wang23]. However, an MRSA ST398 strain from urinary tract infection in a child only carried fnbpB [Reference Monecke24]. Further studies are needed to understand the pathogenic mechanisms of FnBPB in the FnBPB-positive strains isolated from rabbits.
The 281 isolates in this study were only grouped into two STs, ST121 and ST398. Surprisingly, the ST121 strains, but not the ST398 strains, were also detected in a tertiary referral hospital in Xiamen city of Fujian Province [Reference Yu25]. Compared to previous reports, strains belonged to other STs were also isolated in rabbits from Spanish, Thailand and Iberian Peninsula [Reference Viana26–Reference Moreno-Grúa28], indicating that the STs of S. aureus in rabbits might vary geographically. S. aureus strains that belong to ST121 and ST398 were worldwide distributed, and have broad host spectra including both humans and animals. Commonly, ST121 isolates are MSSA that are susceptible to methicillin [Reference Moreno-Grúa28, Reference Kurt29]. In contrast to ST121, strains of ST398 could be further grouped into healthcare-associated, community-associated and livestock-associated strains [Reference David and Daum1, Reference Uhlemann2, Reference Aires-de-Sousa5, Reference Angen6]. Among them, the livestock-associated MRSA ST398 was highly concerned for the transmission to humans from animals [Reference Aires-de-Sousa5]. Transmission of livestock-associated MRSA from swine and bovine to humans had been documented previously [Reference Angen6, Reference van Cleef30]. The case of livestock-associated MRSA ST398 occurring in rabbits and involving farm workers or their family members was also reported in a rabbit farm in Italy [Reference Agnoletti31]. According to previous reports, S. aureus frequently acquired antimicrobial resistance [Reference Uhlemann2–Reference Fu4]. Although isolates resistant to ⩾3 antibiotics were detected in this study, the isolates were all MSSA and no MRSA was detected.
Fujian Province was recognised as one of the key areas for rabbit breeding in China. With the rapid development of rabbit farming in Fujian Province, more studies are needed to understand the epidemiology and characteristics of S. aureus in rabbits and to elucidate its relationship with the human.
Acknowledgements
None.
Financial support
This work was supported by the Outstanding Youth Fund of Fujian Academy of Agricultural Sciences (JC2018-1), Fujian Public Welfare Scientific Research Project (2019R1026-9) and National Rabbit Industry Technology System of People's Republic of China (CARS-43-G-5).
Conflict of interest
None.







