Introduction
Modern medicine and animal husbandry heavily depend on the effectiveness of antimicrobial drugs to combat infectious diseases. The unprecedented rise in resistance to antimicrobial substances throughout the past decades, believed to be linked to their extensive usage in medicine, agriculture, and aquaculture, poses a substantial threat to public health. Indeed, bacterial antimicrobial resistance (AMR) is currently a leading cause of global deaths and is predicted to become one of the greatest public health challenges of the 21st century [Reference Murray, Ikuta, Sharara, Swetschinski, Aguilar, Gray, Han, Bisignano, Rao, Wool, Johnson, Browne, Chipeta, Fell, Hackett, Haines-Woodhouse, Hamadani, Kumaran, McManigal, Achalapong, Agarwal, Akech, Albertson, Amuasi, Andrews, Aravkin, Ashley, Babin, Bailey, Baker, Basnyat, Bekker, Bender, Berkley, Bethou, Bielicki, Boonkasidecha, Bukosia, Carvalheiro, Castañeda-Orjuela, Chansamouth, Chaurasia, Chiurchiù, Chowdhury, Donatien, Cook, Cooper, Cressey, Criollo-Mora, Cunningham, Darboe, Day, Luca, Dokova, Dramowski, Dunachie, Bich, Eckmanns, Eibach, Emami, Feasey, Fisher-Pearson, Forrest, Garcia, Garrett, Gastmeier, Giref, Greer, Gupta, Haller, Haselbeck, Hay, Holm, Hopkins, Hsia, Iregbu, Jacobs, Jarovsky, Javanmardi, Jenney, Khorana, Khusuwan, Kissoon, Kobeissi, Kostyanev, Krapp, Krumkamp, Kumar, Kyu, Lim, Lim, Limmathurotsakul, Loftus, Lunn, Ma, Manoharan, Marks, May, Mayxay, Mturi, Munera-Huertas, Musicha, Musila, Mussi-Pinhata, Naidu, Nakamura, Nanavati, Nangia, Newton, Ngoun, Novotney, Nwakanma, Obiero, Ochoa, Olivas-Martinez, Olliaro, Ooko, Ortiz-Brizuela, Ounchanum, Pak, Paredes, Peleg, Perrone, Phe, Phommasone, Plakkal, Ponce-de-Leon, Raad, Ramdin, Rattanavong, Riddell, Roberts, Robotham, Roca, Rosenthal, Rudd, Russell, Sader, Saengchan, Schnall, Scott, Seekaew, Sharland, Shivamallappa, Sifuentes-Osornio, Simpson, Steenkeste, Stewardson, Stoeva, Tasak, Thaiprakong, Thwaites, Tigoi, Turner, Turner, Doorn, van Velaphi, Vongpradith, Vongsouvath, Vu, Walsh, Walson, Waner, Wangrangsimakul, Wannapinij, Wozniak, Sharma, Yu, Zheng, Sartorius, Lopez, Stergachis, Moore, Dolecek and Naghavi1].
AMR can be transferred between bacteria species via mobile genetic elements, which leads to crossovers between pathogens and commensals in humans, animals, and the environment [Reference von Wintersdorff, Penders, van Niekerk, Mills, Majumder, van Alphen, Savelkoul and Wolffs2], thus making AMR a prime example of a global ‘One Health’ issue [Reference Robinson, Bu, Carrique-Mas, Fèvre, Gilbert, Grace, Hay, Jiwakanon, Kakkar, Kariuki, Laxminarayan, Lubroth, Magnusson, Thi Ngoc, Van Boeckel and Woolhouse3]. While antimicrobial usage (AMU) and AMR in animals are known drivers of human AMR [4, Reference Lazarus, Paterson, Mollinger and Rogers5], the extent to which animals are responsible for AMR in humans, as well as which specific animal sources are most relevant for AMR transmission to humans, is not clear.
Source attribution studies and risk assessments are well suited to investigate the importance of different sources of human AMR [Reference Pires, Duarte and Hald6]. The main source attribution methods are microbial subtyping, comparative exposure assessments, and epidemiological approaches as have been described in detail elsewhere [Reference Mughini-Gras, Kooh, Fravalo, Augustin, Guillier, David, Thébault, Carlin, Leclercq, Jourdan-Da-Silva, Pavio, Villena, Sanaa and Watier7]. Briefly, microbial subtyping studies typically attribute human infections based either on the frequency of source-specific bacteria subtypes in human samples or on the genetic relatedness between human and source strains [Reference Mughini-Gras, Kooh, Fravalo, Augustin, Guillier, David, Thébault, Carlin, Leclercq, Jourdan-Da-Silva, Pavio, Villena, Sanaa and Watier7]. More recently, machine-learning methods, such as random forests, have been applied to, for example, process whole genome sequencing data [Reference Munck, Njage, Leekitcharoenphon, Litrup and Hald8]. Comparative exposure assessments determine the relative importance of different sources of human exposure. Epidemiological approaches include investigations of outbreaks, which summarise source information for multiple outbreaks, and meta-analyses of case–control studies of sporadic cases [Reference Pires, Duarte and Hald6, Reference Mughini-Gras, Kooh, Fravalo, Augustin, Guillier, David, Thébault, Carlin, Leclercq, Jourdan-Da-Silva, Pavio, Villena, Sanaa and Watier7].
Risk assessments are part of risk analysis and can be qualitative, quantitative, or semi-quantitative. For microbial risks, they traditionally follow ‘farm-to-fork’ approaches, which usually combine a hazard description, an assessment of the relationship between hazard and outcome, and an appraisal of the likelihood and magnitude of exposure to the hazard during different phases along the farm-to-fork continuum into a human health risk estimate [Reference Vose, Acar, Anthony, Franklin, Gupta, Nicholls, Tamura, Thompson, Threlfall, Van Vuuren, White, Wegener and Costarrica9]. There are also less data-demanding risk assessment approaches, which, for example, estimate the contribution of a specific source to the total number of human cases, starting at human surveillance data (e.g. [Reference Williams, Ebel and Vose10, Reference Vose, Hollinger and Bartholomew11]).
Objective of the review
A better understanding of the relative contribution of animal sources to human AMR is crucial for planning effective AMR mitigation and control strategies in animals. It is also necessary for an accurate estimation of the economic and public health burden posed by animal diseases, which the Global Burden of Animal Disease study (GBADs), in the context of which this review is undertaken, seeks to produce.
This systematic review aims to summarise the current evidence on the relative contribution of animals or animal products to human AMR, focusing on the methodologies applied to address this question, the investigated antimicrobial hazards, and the included animal sources.
Methods
The completed Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Checklist [Reference Page, McKenzie, Bossuyt, Boutron, Hoffmann, Mulrow, Shamseer, Tetzlaff, Akl, Brennan, Chou, Glanville, Grimshaw, Hróbjartsson, Lalu, Li, Loder, Mayo-Wilson, McDonald, McGuinness, Stewar, Thomas, Tricco, Welch, Whiting and Moher12], as well as additional information about the search strategy, protocol, study selection, data extraction, and compilation of study results, can be found in Supplementary material S1.
Definitions
Antimicrobial resistance describes the ability of microorganisms to survive and uphold pathogenic properties when treated with substances previously effective in eliminating them. It includes resistance to antibiotics, antivirals, antifungals, and antiparasitic agents [13]. This article addresses only the most extensively studied form of AMR, that is, bacterial resistance, and this abbreviation is henceforth used synonymously.
The term ‘animals’ is used to refer to domesticated animals, either for food production (livestock or aquaculture) or companionship (pets). Wildlife is excluded as it is typically studied as a marker for resistance in the environment [Reference Plaza-Rodríguez, Alt, Grobbel, Hammerl, Irrgang, Szabo, Stingl, Schuh, Wiehle, Pfefferkorn, Naumann, Kaesbohrer and Tenhagen14]. We define ‘animal products’ as food items including meat, poultry, fish, shellfish, other aquatic animals, dairy, and eggs [Reference Richardson, Bazaco, Parker, Dewey-Mattia, Golden, Jones, Klontz, Travis, Kufel and Cole15].
Data sources and search strategy
We searched four bibliographic databases (PubMed, Scopus, Embase, and Web of Science) with terms relating to the following key areas: 1) animals and animal products, 2) AMR, 3) relevant study types, and 4) humans. No publication date or language restrictions were applied, but all searches were conducted in English.
Eligibility criteria
We included studies on AMR transmitted from animals or animal products to humans. Studies focusing exclusively on non-human populations, exposure via wild animals or the environment, or other hazards, such as antibiotic agents (i.e., AMU in humans or animals or environmental antibiotic residues) or other types of resistance, were excluded.
Publications were included if they either 1) estimated the number of antibiotic-resistant human infections directly attributable to an animal source, or 2) assessed the relative contribution of at least one animal source to the burden of AMR in humans, either qualitatively or quantitatively. Eligible study types entailed source attribution studies, risk assessments, and any other meeting these criteria. Investigations of outbreaks were included only if they described five or more outbreaks. Those excluded were non-comparative exposure assessments, unrelated studies accidentally picked up by the search string, case studies, case series, single outbreak reports, conference abstracts, studies without new information (reviews, duplicates, or letters to the editors), and studies for which no full text was available. If two or more publications used the same data to illustrate different methodologies, all were included.
Data screening, selection, and extraction
Data screening was conducted in two steps: first, the titles and abstracts were examined based on the inclusion and exclusion criteria, and second, the full texts of the articles included during the first step were retrieved for the final selection. Additionally, we screened the reference lists of the included articles for studies fitting our research objectives and added them to the full-text screening (snowballing).
We defined a data extraction template in Excel and piloted it with a subset of included studies. The final data extraction table (Supplementary material S2) included variables relating to the publication, that is, the study setting, outcome, hazard, methods, and the conceptual approach. The software Sysrev [Reference Bozada, Borden, Workman, Del Cid, Malinowski and Luechtefeld16] was used to track the study selection process, and citations were managed in Zotero [17]. Duplicate entries were removed in Rayyan.ai [Reference Ouzzani, Hammady, Fedorowicz and Elmagarmid18], as Sysrev currently does not offer this function.
Compiling the results of the studies
Due to the broad spectrum of methodologies, study designs, and hazards included, we did not assess the risk of bias or quality of the studies, and it was not possible jointly to analyse the results of the studies. However, to illustrate the relative importance of the animal sources in relation to each other, we assigned ranks according to the results reported by studies that included different animal sources, as described in Supplementary material S1. We also report the most important source for studies on animal and non-animal sources.
The figures were created using Microsoft Office.
Results
Literature review
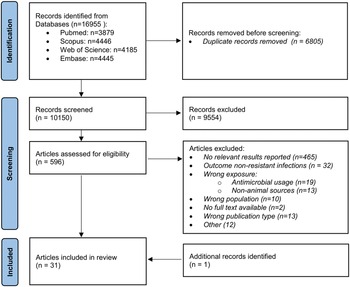
As shown in Figure 1, 16,955 records were initially identified through database searches, 10,150 of which were non-duplicates and screened for eligibility. Of the 596 records selected for full-text screening, 30 articles were included; an additional relevant publication was retrieved via snowballing, thus totalling 31 included studies (Table 1).

Figure 1. Flow chart of included studies.
Country of origin, publication year, and study type
Of the 31 articles, two assessed the impact of different sources on human AMR at a regional level: one for Europe [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19] and the other for South-East Asia [Reference Chereau, Opatowski, Tourdjman and Vong20]. One article provided a framework for any high- or low-income country in South or South-East Asia (SSEA) [Reference Opatowski, Opatowski, Vong and Temime21]. Twenty-four studies were designed in the context of specific countries [Reference Lechner, Freivogel, Stärk and Visschers22–Reference Alban, Olsen, Nielsen, Sørensen and Jessen44], and four were sub-national [Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45–Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48]. Most national and sub-national studies were conducted in North American and European countries, but three studies originated from SSEA countries [Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47]. The highest number (10) of national study publications were from the United States [Reference Vose, Hollinger and Bartholomew11, Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25–Reference Holmberg, Wells and Cohen29, Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32, Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39, Reference Cox and Popken42].
The articles were published over a range of nearly 40 years, mainly as a consequence of a 1984 investigation of resistant salmonellosis outbreaks in the United States [Reference Holmberg, Wells and Cohen29]. However, most studies (87%) were published between 2012 and 2022, and 12 have been published since 2020 [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Opatowski, Opatowski, Vong and Temime21, Reference Lechner, Freivogel, Stärk and Visschers22, Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25, Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30, Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34, Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35, Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38–Reference Collineau40, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47].
Based on the authors’ descriptions and reported methods, 16 articles were categorised as source attribution studies, including seven microbial subtyping studies [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30–Reference Hald, Lo Fo Wong and Aarestrup33, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47], five investigations of outbreaks [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25–Reference Holmberg, Wells and Cohen29], three comparative exposure assessments [Reference Lechner, Freivogel, Stärk and Visschers22–Reference Carmo, Nielsen, da Costa and Alban24], and one multi-directional dynamic risk model [Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34]. Twelve were classified as risk assessments [Reference Vose, Hollinger and Bartholomew11, Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Opatowski, Opatowski, Vong and Temime21, Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38–Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48], and the remaining three were of a different study type [Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35–Reference Bosch, van Luit, Pluister, Frentz, Haenen, Landman, Witteveen, van Marm-Wattimena, van der Heide and Schouls37].
Methodological frameworks
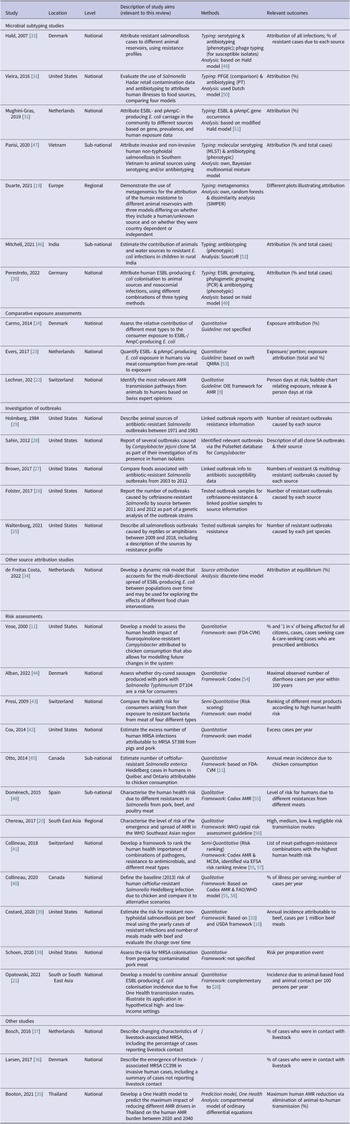
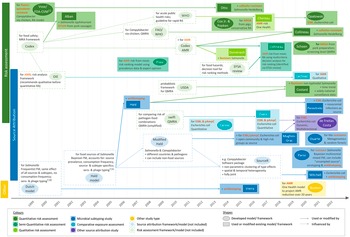
Table 1 summarises the countries of origin, relevant aims, methods, and outcome measures of the articles by study type. Figure 2 shows their timeline and the methods and guidelines they were based on to illustrate how they developed over time.
Table 1. Characteristics of the included studies

Abbreviations: Codex, Codex Alimentarius; EFSA, The European Food Safety Authority; ESBL, extended-spectrum β-lactamase; FAO, Food and Agriculture Organization; FDA-VCM, Food and Drug Administration Center for Veterinary Medicine; MRSA, methicillin-resistant S. aureus; OIE, World Organisation for Animal Health (now WOAH); (p)AmpC, (plasmid)-mediated AmpC β-lactamase; QMRA, quantitative microbial risk assessment; USDA, United States Department of Agriculture; WHO, World Health Organization.

Figure 2. Timeline of the studies and links between their methodologies (excluding investigations of outbreaks). Words printed in red describe how the study addressed AMR. AMR, bacterial antimicrobial resistance; EFSA, The European Food Safety Authority; ESBL, extended-spectrum ß-lactamase producing; FAO, Food and Agriculture Organization; FDA-VCM, Food and Drug Administration Center for Veterinary Medicine; FM, frequency matched model; MRSA, methicillin-resistant Staphylococcus aureus; (p)AmpC, (plasmid)-mediated AmpC ß-lactamase producing; RA, risk assessment; (Q)MRA, (quantitative) microbial RA; OIE, World Organisation for Animal Health (now WOAH); WHO, World Health Organization.
Microbial subtyping
Of the seven microbial subtyping studies, one used metagenomics and random forests to attribute the entirety of resistance genes, and the human resistome, to different sources [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19]. The rest followed frequency-matched approaches. A study on resistant Salmonella Hadar infections used a well-known frequentist model [Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32], also called the ‘Dutch model’ [Reference van Pelt, Giessen and Leeuwen50]. One study on Salmonella spp. [Reference Hald, Lo Fo Wong and Aarestrup33] and three on different Escherichia coli strains [Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30, Reference Mughini-Gras, Dorado-García, van Duijkeren, van den Bunt, Dierikx, Bonten, Bootsma, Schmitt, Hald, Evers, de Koeijer, van Pelt, Franz, Mevius and Heederik31, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46] were based on a Bayesian subtyping model, the ‘Hald model’ [Reference Hald, Vose, Wegener and Koupeev49], its modification [Reference Mullner, Jones, Noble, Spencer, Hathaway and French51], or SourceR for quantitative source attribution [Reference Miller, Marshall, French and Jewell52]. Finally, Parisi and colleagues developed a Bayesian multinomial mixed model for their study on non-typhoidal salmonellosis [Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47]. More details about the models and adaptations made by the individual studies can be found in Figure 2.
Three microbial subtyping studies compared the results of different combinations of subtyping methods [Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30, Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47], and two assessed the impact of including ‘human-to-human transmission’ or an ‘unspecified’ source [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30]. Duarte and colleagues further defined country-dependent and country-independent models for the data they collected from several European countries [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19]. The studies generally found that the more subtyping approaches were combined, the less likely that cases were attributed to specific animal sources and the more relevant unknown or human sources became.
Epidemiological approaches
Five studies from the United States reported on animal sources of outbreaks caused by resistant infections. For two of them, both on salmonellosis, the assessment of the relative contributions of the different sources was the main study aim [Reference Brown, Grass, Richardson, Nisler, Bicknese and Gould27, Reference Holmberg, Wells and Cohen29]. Two studies included the sources of resistant outbreaks as part of answering different research questions: one concerned Campylobacter jejuni clone SA, a tetracycline-resistant strain [Reference Sahin, Fitzgerald, Stroika, Zhao, Sippy, Kwan, Plummer, Han, Yaeger and Zhang28], and the other ceftriaxone-resistant Salmonella [Reference Folster, Grass, Bicknese, Taylor, Friedman and Whichard26]. The fifth study reported the relative importance of different reptilian or amphibian pet species in causing antibiotic-resistant salmonellosis outbreaks [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25]. We did not identify any meta-analyses on sporadic resistant infections in humans.
Comparative exposure assessments
Two of the three comparative exposure assessments identified were quantitative and focused on the relative contribution of different meat types to human exposure by antibiotic-resistant E. coli strains [Reference Evers, Pielaat, Smid, van Duijkeren, Vennemann, Wijnands and Chardon23, Reference Carmo, Nielsen, da Costa and Alban24]. The third study qualitatively assessed the relative importance of AMR exposure via different animal sources [Reference Lechner, Freivogel, Stärk and Visschers22]. The frameworks of the studies are described in Figure 2.
Lastly, a study was identified which attributed extended-spectrum β-lactamase (ESBL)-producing E. coli infections in the community to different sources using a dynamic risk model that allows for multi-directional pathogen transmission between human and animal populations over time [Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34].
Risk assessments
Of the 12 risk assessments, seven were quantitative [Reference Vose, Hollinger and Bartholomew11, Reference Opatowski, Opatowski, Vong and Temime21, Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39, Reference Collineau41, Reference Cox and Popken42, Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45], two were semi-quantitative [Reference Collineau41, Reference Presi43], two were qualitative [Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48], and one combined qualitative with quantitative parts [Reference Alban, Olsen, Nielsen, Sørensen and Jessen44]. Seven studies either followed the Codex Alimentarius guidelines for microbial food safety hazards risk assessments (Codex) [54, 55], or frameworks based on, or influenced by, them [Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Opatowski, Opatowski, Vong and Temime21, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39–Reference Collineau41, Reference Alban, Olsen, Nielsen, Sørensen and Jessen44, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48]. Another study did not specify any guidelines, but was also in line with the overall Codex framework [Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38]. Three risk assessments developed their own models [Reference Vose, Hollinger and Bartholomew11, Reference Cox and Popken42, Reference Presi43]. One of them, the Food and Drug Administration Center for Veterinary Medicine (FDA-CVM) guideline [Reference Vose, Hollinger and Bartholomew11], was applied by a separate Canadian study [Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45]; see Figure 2.
Other studies
Three studies did not fit into the above-described categories, namely: a One Health prediction model of the maximum future human health impact of eliminating different AMR transmission routes in Thailand [Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35] and two studies describing the proportion of cases of livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) in the Netherlands [Reference Bosch, van Luit, Pluister, Frentz, Haenen, Landman, Witteveen, van Marm-Wattimena, van der Heide and Schouls37] and Denmark [Reference Larsen, Petersen, Larsen, Sieber, Stegger, Koch, Aarestrup, Price, Skov, Johansen, Westh, Pedersen, Jensen, Jensen, Chen, Strøbæk, Østergaard, Lomborg, Ellermann-Eriksen and Ripadal36] among individuals who had livestock contact previous to their infection. While the latter two studies technically met the criteria for inclusion, they did not utilise any specific methodology to address our research objective and, therefore, will not be addressed further.
Conceptual approaches
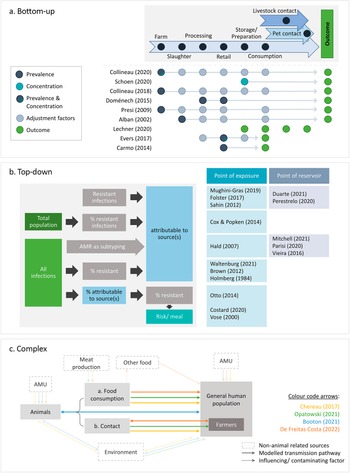
The articles differed in their conceptual approaches for modelling the link between animal sources and human AMR. All comparative exposure assessments [Reference Lechner, Freivogel, Stärk and Visschers22–Reference Carmo, Nielsen, da Costa and Alban24] and six risk assessments [Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38, Reference Collineau40, Reference Collineau41, Reference Presi43, Reference Alban, Olsen, Nielsen, Sørensen and Jessen44, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48] followed a bottom-up approach, starting at the hazard at one point of the food production chain and factoring in the effects of subsequent steps to estimate the human health impact at exposure (Figure 3a). In addition to food-related exposure, Lechner and colleagues included direct animal contact as an exposure route [Reference Lechner, Freivogel, Stärk and Visschers22].

Figure 3. Conceptual approaches of the identified studies. (a) Bottom-up approaches: start at one point along the farm-to-fork continuum and adjust for different factors to arrive at an estimate of the health outcome. (b) Top-down approaches: start at the outcome and attribute it to one or multiple sources, either at the level of exposure (i.e., consumption or contact) or at the animal reservoir. (c) Complex approaches: integrate all One Health domains or account for multi-directional hazard transmission. Arrows indicate the directionality of transmission. AMR, bacterial antimicrobial resistance; AMU, antimicrobial usage.
Sixteen studies started at the outcome and either estimated the contribution of one specific source to it [Reference Vose, Hollinger and Bartholomew11, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39, Reference Cox and Popken42, Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45] or attributed it to different sources [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25–Reference Hald, Lo Fo Wong and Aarestrup33, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47]. All microbial subtyping studies [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30–Reference Hald, Lo Fo Wong and Aarestrup33, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47], all investigations of outbreaks [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25–Reference Holmberg, Wells and Cohen29], and four risk assessments [Reference Vose, Hollinger and Bartholomew11, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39, Reference Cox and Popken42, Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45] adopted such ‘top-down’ concepts (Figure 3a). Eleven studies attributed infections at the point of consumption or contact (exposure), and the remaining five partitioned the outcome to animal reservoirs. The latter included one quantitative risk assessment [Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39] and all microbial subtyping studies, except Mughini-Gras et al. [Reference Mughini-Gras, Dorado-García, van Duijkeren, van den Bunt, Dierikx, Bonten, Bootsma, Schmitt, Hald, Evers, de Koeijer, van Pelt, Franz, Mevius and Heederik31] and Hald et al. [Reference Hald, Lo Fo Wong and Aarestrup33], both of which integrated information on the exposure frequency to attribute infections at the point of exposure.
Four studies employed more complex concepts that did not fit into the top-down or bottom-up approaches (Figure 3c). Two of these followed a One Health strategy [Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Opatowski, Opatowski, Vong and Temime21], another allowed for multi-directional AMR transfer between animals and humans [Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34], and the last had both a One Health concept and addressed multi-directional spread between populations [Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35].
Hazard and outcome definitions
Most studies investigated infections due to specific bacteria–resistance combinations. Three articles reported on bacterial subtypes known to be associated with specific resistances [Reference Sahin, Fitzgerald, Stroika, Zhao, Sippy, Kwan, Plummer, Han, Yaeger and Zhang28, Reference Cox and Popken42, Reference Alban, Olsen, Nielsen, Sørensen and Jessen44], and five focused on general resistance in a specific pathogen [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25, Reference Brown, Grass, Richardson, Nisler, Bicknese and Gould27, Reference Holmberg, Wells and Cohen29, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48] (Figure 4a). Four microbial subtyping studies included all infections with the pathogen and accounted for resistance by using antibiotic profiles to subtype strain populations [Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32, Reference Hald, Lo Fo Wong and Aarestrup33, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47]. Opatowski and colleagues built a generic quantitative model applicable for any resistant pathogen [Reference Opatowski, Opatowski, Vong and Temime21], whereas four other articles considered multiple bacteria–resistance combinations relevant to human health [Reference Collineau41, Reference Presi43] or qualitatively addressed AMR in general [Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Lechner, Freivogel, Stärk and Visschers22]. The only quantitative assessment that focused on the resistome as a whole was that by Duarte and colleagues [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19].
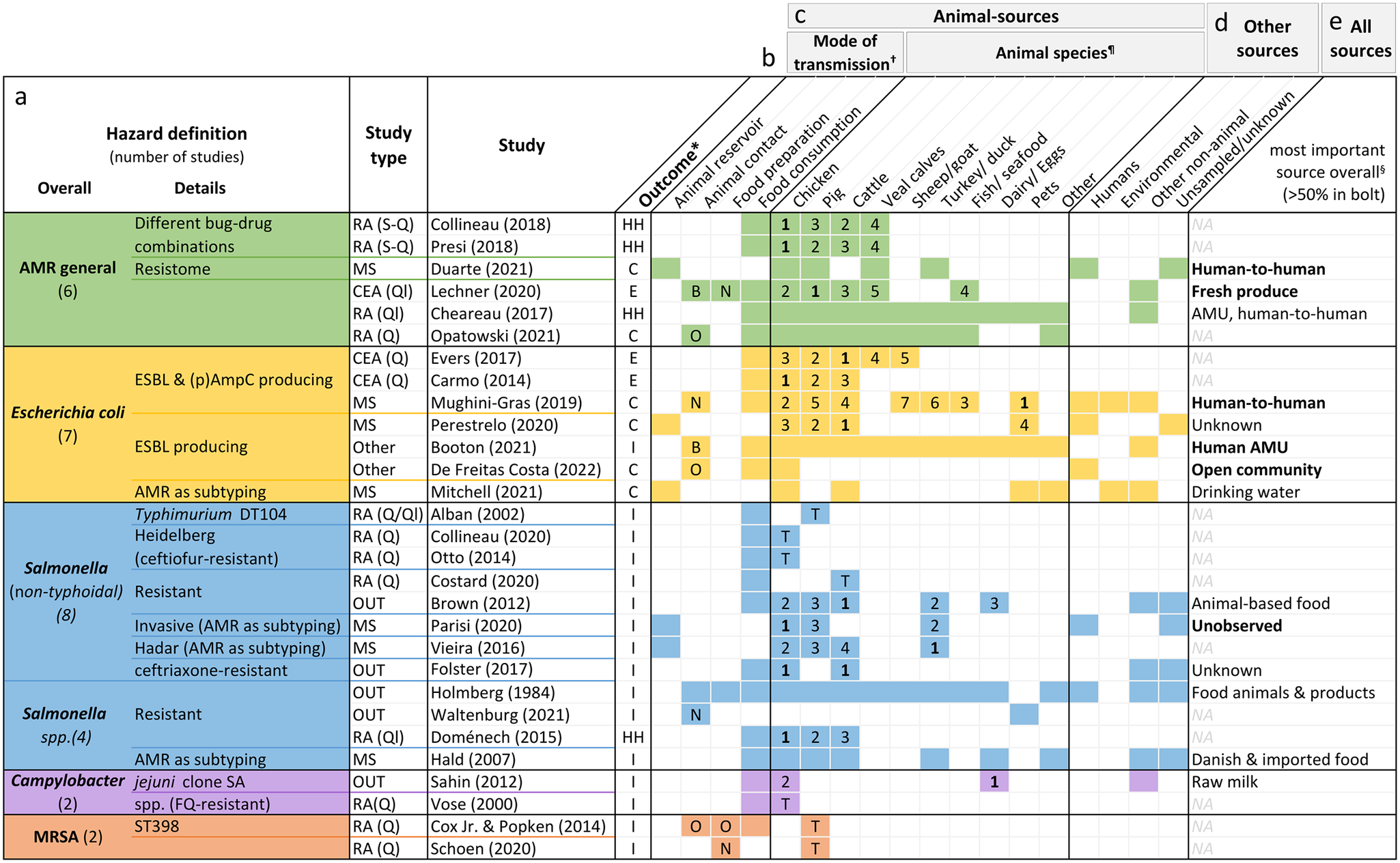
Figure 4. Hazard definition (a), outcome measure (b), investigated animal-related (c) and non-animal-related sources (d), as well as the most important source found (e) by the included studies. Colourised fields indicate that the respective source(s) and mode(s) of transmission were addressed by the study.
*Outcomes: overall human health risk due to hazard (HH), human colonisation with hazard (H), human illness due to hazard (I), or exposure to hazard (E).
†Letters indicate whether food preparation and animal contact were occupational (O), non-occupational (N) or both (B). If no letter, it was not specified.
¶The numbers are ranks of importance (1 = highest importance). They are only given for studies reporting the relative contribution of different animal sources to human AMR in relation to each other. Studies examining risk due to only one specific animal source are shown with a T (total risk estimate).
§Only given for studies with both animal and non-animal sources. If the source is printed in bold, the study found it to be responsible for over 50% of the outcome. AMR, bacterial antimicrobial resistance; AMU, antimicrobial usage; CEA, comparative exposure assessment; DT, definite/phage type; ESBL, extended-spectrum ß-lactamase; FQ, fluoroquinolone; MRSA, methicillin-resistant Staphylococcus aureus; NA, not applicable; OUT, investigation of outbreaks; (p)AmpC, (plasmid)-mediated AmpC ß-lactamase; RA, risk assessment; S-Q, semi-quantitative; Q, quantitative; Ql, qualitative.
Of the articles on specific pathogens, 12 concerned Salmonella species. Seven investigated E. coli, mainly strains producing ESBL and/or (plasmid)-mediated AmpC β-lactamase. Two studies each were about Campylobacter and MRSA.
Sixteen studies defined the outcome as human illness due to resistant pathogens, six investigated colonisation with resistant bacteria, and all four qualitative and semi-quantitative risk assessments defined it as a health risk caused by AMR. The key outcome of the three exposure assessments was exposure to AMR (Figure 4b).
Animal sources
Five studies investigated animal reservoirs [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19, Reference Perestrelo, Correia Carreira, Valentin, Fischer, Pfeifer, Werner, Schmiedel, Falgenhauer, Imirzalioglu, Chakraborty and Käsbohrer30, Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32, Reference Mitchell, Purohit, Jewell, Read, Marrone, Diwan and Stålsby Lundborg46, Reference Parisi, Phuong, Mather, Jombart, Tuyen, Lan, Trang, Carrique-Mas, Campbell, Trung, Glass, Kirk and Baker47], 15 focused on food consumption [Reference Vose, Hollinger and Bartholomew11, Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Evers, Pielaat, Smid, van Duijkeren, Vennemann, Wijnands and Chardon23, Reference Carmo, Nielsen, da Costa and Alban24, Reference Folster, Grass, Bicknese, Taylor, Friedman and Whichard26–Reference Sahin, Fitzgerald, Stroika, Zhao, Sippy, Kwan, Plummer, Han, Yaeger and Zhang28, Reference Hald, Lo Fo Wong and Aarestrup33, Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39–Reference Collineau41, Reference Presi43–Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45, Reference Doménech, Jiménez-Belenguer, Pérez, Ferrús and Escriche48], one focused on animal contact [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25], one focused on food preparation [Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38], and seven included different manners of acquisition [Reference Opatowski, Opatowski, Vong and Temime21, Reference Lechner, Freivogel, Stärk and Visschers22, Reference Holmberg, Wells and Cohen29, Reference Mughini-Gras, Dorado-García, van Duijkeren, van den Bunt, Dierikx, Bonten, Bootsma, Schmitt, Hald, Evers, de Koeijer, van Pelt, Franz, Mevius and Heederik31, Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34, Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35, Reference Cox and Popken42] (Figure 4c).
Chicken was the most investigated animal source with 20 studies, followed by cattle (including veal calves) with 19 studies and pigs with 16 studies. Other animal sources included turkeys (n = 5), sheep or goat (n = 2), fish or seafood (n = 2), eggs (n = 2), raw milk (n = 1), and duck (n = 1) (Figure 4c). Four articles explored the role of pets as sources of AMR, one of which examined specifically the relative importance of pet reptiles or amphibians in causing resistant outbreaks [Reference Waltenburg, Perez, Salah, Karp, Whichard, Tolar, Gollarza, Koski, Blackstock, Basler and Nichols25].
Of the risk assessments estimating the human risk due to a single animal source of AMR, three concerned chicken products [Reference Vose, Hollinger and Bartholomew11, Reference Collineau41, Reference Otto, Carson, Finley, Thomas, Reid-Smith and McEwen45], three pig products [Reference Schoen, Peckham, Shirai, Kissel, Thapaliya, Smith and Meschke38, Reference Cox and Popken42, Reference Alban, Olsen, Nielsen, Sørensen and Jessen44], and one beef [Reference Costard, Pouzou, Belk, Morley, Schmidt, Wheeler, Arthur and Zagmutt39]. Fourteen studies also included non-animal-related sources, such as human-to-human exposure or environmental sources (Figure 4d).
Resulting estimates
The studies reported their results in one or more of the following ways: 1) as the relative contribution of different animal-related and non-animal sources to the human AMR burden (percentage, total numbers, or qualitatively), 2) as the relative importance of animal sources in relation to each other, or 3) as the total number of human cases due to a specific animal source (per population or per meal). Descriptions of the results reported by each study can be found in Table 1.
Figure 4e lists the most important contributors to the human AMR burden found by studies with outcome type 1. The estimates differed with the pathogen under investigation; while none of the five studies on resistant E. coli found animals to be the most important contributor, most studies on resistant Salmonella attributed the highest proportion of illness to animal sources.
In Figure 4c, the results of studies reporting outcomes of the second type are displayed in the form of ranks. Chicken was most frequently assigned the top rank (six times), followed by cattle which was most relevant according to four studies. One study found turkeys to be the major contributor [Reference Vieira, Grass, Fedorka-Cray, Plumblee, Tate and Cole32] while in another study pets were more relevant than individual livestock species – but not when compared to all livestock species combined [Reference Mughini-Gras, Dorado-García, van Duijkeren, van den Bunt, Dierikx, Bonten, Bootsma, Schmitt, Hald, Evers, de Koeijer, van Pelt, Franz, Mevius and Heederik31].
Discussion
We aimed to describe the current state of evidence on the relative direct contribution of animal sources to human AMR. Literature searches were kept broad to capture as many relevant publications as possible and screened over 10,000 records; however, we only found 31 studies that addressed our specific research objective. This illustrates that while there is a large and growing body of evidence on the topic of AMR, there is still a paucity of studies compiling this evidence to assess the concrete contribution of animals to human AMR.
Our work complements a 2018 review by Pires and colleagues, which describes the utility of risk assessments and source attribution studies for determining the relative importance of different sources of human AMR [Reference Pires, Duarte and Hald6]. We systematically searched for publications following such or similar strategies, albeit focusing on animal sources of AMR. Our results support their observation of these study types still being in their infancy in the context of AMR [Reference Pires, Duarte and Hald6]. However, there has been a marked recent increase of relevant publications as almost 40% of studies were published in the past three years alone. This rise coincided with the publication of the Global Action Plan on Antimicrobial Resistance (GAP) by WHO in 2015, which may have contributed to the momentum of AMR research [59].
Most microbial subtyping studies were published in Europe, which was unsurprising, given that this study type originated in Denmark and the Netherlands [Reference Hald, Vose, Wegener and Koupeev49, Reference van Pelt, Giessen and Leeuwen50]. North America contributed a third of the studies, including all five investigations of outbreaks, which were all from the United States. The availability of nationwide surveillance systems such as the National Antimicrobial Resistance Monitoring System (NARMS) [60], the CDC’s Foodborne Disease Active Surveillance Network (FoodNet) [61], and the National Outbreak Reporting System (NORS) [62] are likely among the reasons for why we identified multiple US studies compiling source information for resistant outbreaks. However, similar systems also exist in other countries, for example the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) [63] and FoodNet Canada [64]. The lack of comparable outbreak studies from other countries may, therefore, not reflect a true absence of evidence, but could also be influenced by the limitations of our search scope.
We did not find any studies from South America, Africa, or Oceania, which may indicate either that study types relevant to this review are not published in scientific journals in these regions or that no such studies are conducted there, possibly due to data scarcity or lack of awareness or interest in the issue.
Given the nature of the AMR hazard, capturing the complete burden it causes in humans via animal sources requires approaches accounting for horizontal gene transfer. However, most studies focused on specific pathogens, which reflects the way infectious disease data are currently collected. Only one study quantitatively investigated sources of the human resistome [Reference Duarte, Röder, Van Gompel, Petersen, Hansen, Hansen, Bossers, Aarestrup, Wagenaar and Hald19]. Increasing usage of (meta-)genomics for resistance characterisation in food safety in recent years, combined with decreasing sequencing costs [Reference Pennone, Cobo-Díaz, Prieto and Alvarez-Ordóñez65], gives hope that the availability of data necessary to conduct similar studies will increase over the coming years.
A commonly stated limitation of the source attribution studies was their inability to account for bidirectional AMR transmission between animals and humans. However, only two studies identified explicitly integrated directionality into their models: one developing a multi-directional dynamic risk model [Reference de Freitas Costa, Hagenaars, Dame-Korevaar, Brouwer and de Vos34] and the other specifying a human-to-animal transmission parameter in their prediction model [Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35].
While our focus was on the direct impact of animal sources on human AMR, we acknowledge that AMR is an issue spanning across animals, humans, and the environment and that more holistic approaches are necessary to describe fully all direct and indirect connections between them. Indeed, we found three studies that followed such One Health concepts [Reference Chereau, Opatowski, Tourdjman and Vong20, Reference Opatowski, Opatowski, Vong and Temime21, Reference Booton, Meeyai, Alhusein, Buller, Feil, Lambert, Mongkolsuk, Pitchforth, Reyher, Sakcamduang, Satayavivad, Singer, Sringernyuang, Thamlikitkul, Vass, Avison, Alhusein, Booton, Buller, Chantong, Charoenlap, Couto, Dulyayangkul, Feil, Gibbon, Gould, Lambert, Meeyai, Mongkolsuk, Montrivade, Pitchforth, Phoonsawad, Rangkadilok, Ratanakorn, Reyher, Sakcamduang, Satayavivad, Singer, Sirikanchana, Sringernyuang, Suriyo, Suwanpakdee, Thamlikitkul, Turner, Vass, Wichuwaranan, Wiratsudakul, Avison and Turner35].
None of the studies included subgroup analyses for vulnerable populations. Gender or socioeconomic factors were also not investigated. Such factors may likely have a considerable impact on the probability of AMR acquisition [Reference Forrester, Cao, Schaps, Liou, Patil, Stave, Sokolow and Leo66, Reference Jones, Mitchell, Cooke, Baral, Arjyal, Shrestha and King67] and should be kept in mind for future research.
Fish and seafood were only addressed by two studies. Given the rapid growth of the aquaculture sector and the accompanying use of antimicrobials, especially in regions without adequate regulations, pathogens of aquatic origins have high resistance proportions [Reference Watts, Schreier, Lanska and Hale68]. Aquaculture, therefore, presents a potential threat to human health and merits further research.
We compiled some results of the studies to give a broad overview, but when interpreting them, it should be kept in mind that no account was made for study design, quality, or uncertainty and the results were merely ranked for each study individually. The findings suggest differing degrees of importance of the animal sources depending on the type of hazard, which is in line with other studies observing different risks of exposure via the same type of meat source depending on the bacteria–resistance combination [Reference Zhang, Schmidt, Arthur, Wheeler and Wang69, Reference Plaza-Rodríguez, Mesa-Varona, Alt, Grobbel, Tenhagen and Kaesbohrer70].
By aiming at identifying studies that quantified the relative contribution of animal sources to human AMR, we implicitly posed the underlying assumption that there is a link between animal and human AMR. The question of whether transmission of resistance occurs between animals and humans has been addressed by previous systematic reviews. For example, such a review from 2018 found that 33 out of 45 eligible studies supported the view that transmission of resistant E. coli occurs between food-producing animals and humans [Reference Muloi, Ward, Pedersen, Fèvre, Woolhouse and van Bunnik71]. Likewise, another systematic review summarised information on food-producing animals as potential origins for extra-intestinal E. coli resistant to cephalosporins and found overall supporting evidence, especially for poultry [Reference Lazarus, Paterson, Mollinger and Rogers5].
Given our focus on the direct contribution of animals to human AMR, we did not include AMU in our search strategy and excluded studies attributing cases of AMR in humans solely to AMU in animals. However, even when omitting AMU-specific search terms, during the screening, we observed several studies, especially risk assessments, relating AMU in animals to human AMR, indicating that there is considerable evidence available on this topic that could be explored by future research.
We did not search grey literature databases and, therefore, likely missed relevant publications, such as government reports. Furthermore, while articles were not excluded based on language, our searches were conducted in English, thereby likely missing relevant publications in other languages.
In conclusion, the body of evidence on the direct contribution of animal sources to AMR in humans is growing but still relatively small. Existing studies utilise a broad range of methodologies to address this question. Recent years have seen promising developments, such as using human resistome data for source attribution, that will aid in tailoring studies to the specific characteristics of the AMR hazard.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268823001309.
Data availability statement
The search strategies and all extracted data can be found in the Supplementary Materials.
Author contribution
Conceptualization: S.M.P., L.M., B.D., C.D.B., J.S.A., S.B.M., N.V., D.P., C.F.; Writing – review & editing: S.M.P., L.M., B.H., B.D., C.D.B., H.C.D.C.F., J.R., J.S.A., S.B.M., N.V., D.P., C.F.; Funding acquisition: B.H., B.D., J.R., D.P.; Methodology: B.D., C.D.B., H.C.D.C.F., J.S.A., S.B.M., N.V., D.P.; Project administration: B.D., D.P.; Supervision: B.D.; Investigation: H.C.D.C.F., C.F.; Validation: H.C.D.C.F., C.F.; Data curation: C.F.; Visualization: C.F.; Writing – original draft: C.F.
Financial support
This research was performed in the framework of the Global Burden of Animal Diseases (GBADs) programme, which is led by the University of Liverpool and the World Organization for Animal Health (WOAH) (https://animalhealthmetrics.org/). This research was supported through the Grant Agreement Investment ID INV-005366 with the Bill & Melinda Gates Foundation and the UK Foreign, Commonwealth and Development Office (FCDO). GBADs case studies received additional funding from the following: European Commission, Australian Centre for International Agricultural Research (ACIAR), Brooke Foundation and the Food and Agriculture Organization of the United Nations (FAO). A full list of the GBADs collaborators can be accessed here: https://animalhealthmetrics.org/acknowledgements. This work was also supported by a Subgrant Agreement (award number 20220419) with Fleming Fund and WOAH.
Competing interest
The authors declare none.