Introduction
Rationale
The influence of human activities, including agriculture, on the amount of antibiotic-resistant bacteria (ARB) or associated antibiotic resistance genes (ARG) in the environment is an area of concern with potential risks for human, animal and ecosystem health (Ashbolt et al., Reference Ashbolt, Amézquita, Backhaus, Borriello, Brandt, Collignon, Coors, Finley, Gaze, Heberer, Lawrence, Larsson, McEwen, Ryan, Schönfeld, Silley, Snape, Van den Eede and Topp2013; Perry and Wright, Reference Perry and Wright2013; Rizzo et al., Reference Rizzo, Manaia, Merlin, Schwartz, Dagot, Ploy, Michael and Fatta-Kassinos2013; Varela and Manaia, Reference Varela and Manaia2013; Larsson, Reference Larsson2014; Singer and Williams-Nguyen, Reference Singer and Williams-Nguyen2014; Williams-Nguyen et al., Reference Williams-Nguyen, Sallach, Bartelt-Hunt, Boxall, Durso, McLain, Singer, Snow and Zilles2016). Numerous studies have detected ARB and ARG in a variety of environmental sites globally, including seawater, fresh water, soil, and air. Although antibiotic resistance in environmental bacteria has existed for billions of years and occurs in locations across the globe, some evidence suggests that the amount of resistance in environmental media increased dramatically in the last century, likely due to the extensive use of pharmaceutical antibiotics (Finley et al., Reference Finley, Collignon, Larsson, McEwen, Li, Gaze, Reid-Smith, Timinouni, Graham and Topp2013). Despite this, the degree to which increased frequency of these resistance factors in the environment are the result of specific anthropogenic sources, such as human wastewater or run-off from animal agriculture, is an area of considerable uncertainty (Wooldridge, Reference Wooldridge2012; Woolhouse et al., Reference Woolhouse, Ward, van Bunnik and Farrar2015). Point sources, such as wastewater effluent pipes and agricultural waste lagoons represent an important and definable contribution to this environmental problem. Here we present a method for summarizing the available scientific evidence pertaining to the effect of point sources on bacterial antibiotic resistance in the environment.
Systematic reviews are a rigorous knowledge synthesis technique used extensively in the health sciences to summarize information from numerous randomized trials examining the clinical efficacy of an intervention (Sargeant and O'Connor, Reference Sargeant and O'Connor2014a, Reference Sargeant and O'Connorb). In contrast to a narrative review, this method permits the evaluation of all available evidence on the question of interest using a standardized process. A documented search strategy gathers all published literature on a predefined question, and identified publications are screened for relevance to this question. For included studies, relevant data are extracted, risk of bias to the internal validity of the study is assessed, and the overall evidence is summarized.
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement, published in 2009, proposed reporting standards for systematic reviews to ensure thorough and consistent reporting of systematic review activities (Moher et al., Reference Moher, Liberati, Tetzlaff and Altman2009). The PRISMA statement and its recent extension, PRISMA for protocols (PRISMA-P) (Moher et al., Reference Moher, Shamseer, Clarke, Ghersi, Liberati, Petticrew, Shekelle and Stewart2015), recommends creating and making accessible a predefined protocol describing how the systematic review will be conducted. Here we describe the protocol for a systematic review of available evidence on the subject of whether environmental levels of bacterial antibiotic resistance are higher in close proximity to point sources of anthropogenic effluent compared to areas more distant from those sources.
Objectives
This protocol describes a method to rigorously evaluate the state of evidence on the following question: Is the prevalence or concentration of antibiotic resistant bacteria or resistance genes in soil, water, air or free-living wildlife higher in close proximity to, downstream from or downwind from, known or suspected sources compared to areas more distant, upstream, or upwind from these sources? The overall goals of this systematic review are to provide comprehensive information about the state of knowledge on this systematic review question and identify gaps in scientific knowledge. The purpose of this protocol is to provide a priori documentation of the methods and process that will be used for the systematic review.
Review team roles and responsibilities
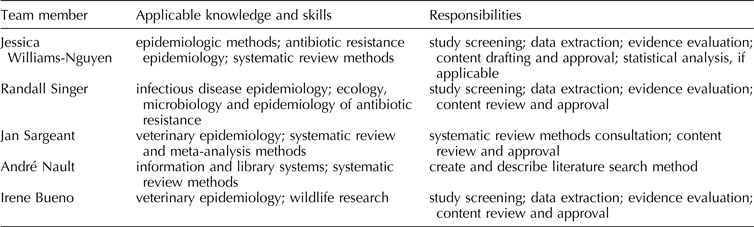
Planned review team members, including information about members’ applicable knowledge, skills, and responsibilities, are listed in Table 1.
Table 1. Planned review team membership
Methods
Eligibility criteria
Criteria for eligibility (Table 2) for this systematic review will be based on the Population, Exposure, Comparator, Outcome, Study design (PECOS) framework. Studies of this question are expected to be predominantly cross-sectional in design based on prior knowledge of the available literature. Randomized trials of this question, by randomized field application of manure for example, will be included if they examine the influence of proximity or direction to these randomized sources on resistance factors. We note that ‘close proximity’ is not defined a priori in this systematic review protocol in order to avoid exclusion of potentially relevant studies. The range of possible distance values examined in the literature is unknown, and there is no commonly accepted standard or cut-off for the distance at which expected effects of point sources on resistance outcomes in environmental media will occur.
Table 2. Eligibility criteria for inclusion of studies in a systematic review on whether point sources of anthropogenic effluent increase antibiotic resistance in the environment

ARB, antibiotic resistant bacteria; ARG, antibiotic resistance genes.
Sources of information
An explicit and comprehensive strategy will be used to search available scientific literature. Searches of the following electronic databases from inception dates without language restrictions will be conducted: PubMed, CABI, and Scopus. The search strategy used for each database will be recorded.
Search strategy
PubMed search strategy will be as follows:
“drug resistance, microbial”[Mesh] AND (“water pollutants”[Mesh] OR “environment”[MeSH Terms] OR “soil”[MeSH Terms] OR “water”[MeSH Terms] OR “water pollution”[MeSH Terms] OR “air pollution”[MeSH Terms] OR “air pollutants”[MeSH Terms] OR “animals, wild”[MeSH Terms]) AND (“Animals”[MeSH Terms] OR “humans”[MeSH Terms] OR “animal feed”[MeSH Terms] OR “manure”[MeSH Terms] OR “aquaculture”[MeSH Terms] OR “waste water”[MeSH Terms] OR “sewage”[MeSH Terms] OR “hospitals”[MeSH Terms] OR “hospitals, animal”[MeSH Terms] OR “cities”[MeSH Terms]) NOT “therapeutics”[MeSH Terms] NOT “drug discovery”[MeSH Terms] NOT “aids”[All Fields] NOT “hiv”[All Fields] NOT “influenza”[All Fields]
Search strategies for Scopus and CABI will be modeled after the strategy above. In addition to the electronic literature searches, hand searches of the references listed in relevant narrative reviews and key research articles will also be conducted to ensure all relevant peer reviewed literature is assessed. Narrative reviews of interest will be identified from search results during relevance screening as well as from the review team's knowledge of the literature.
Data management
The titles and abstracts, when available, of the articles identified by the searches will be downloaded into EndNote (Thomson Reuters), a reference management software package. Screening, quality assessment and data extraction will be recorded in Excel 2013 (Microsoft Corporation, Redmond, WA, USA). Statistical analyses, if conducted, will be done using SAS 9.4 for Windows (2002–2010, SAS Institute Inc., Cary, NC, USA).
Selection of studies
Relevance screening of abstracts will be independently conducted by at least two reviewers to exclude articles that do not address the study question. Consensus between the reviewers on relevance of the article will be required. Any conflicts between reviewers in screening, quality assessment or data extraction decisions will be resolved by phone conference with the review team on a weekly basis.
Relevance screening will consist of the following questions:
-
(1) Does the abstract refer to primary research reported in a journal publication or a thesis (as opposed to a review article or presentation abstract or proceedings)?
-
(2) Were samples of soil, water, or air or biological samples from wildlife species collected from the exterior environment (i.e. not in a building or industrial facility)?
-
(3) Does the study measure the prevalence or concentration of bacterial antibiotic resistance factors (bacteria or genes) in the samples?
Any article for which the answer to at least one of these questions is ‘No’ will be excluded from further consideration without additional review. All articles for which the answer is ‘Yes’ to all relevance screening or for which the answer cannot be determined from the abstract will be determined to be potentially relevant and will be retained for additional screening questions. Aside from thesis documents, book chapters will not be eligible for inclusion.
Full-text of these publications will be subjected to a second level of screening, which we will term design screening. Only the methods section of each publication will be reviewed at this stage. Reviewers will not look at results or conclusions when determining the screening outcome. This screening step will be independently conducted by two reviewers using a standardized questionnaire, and agreement will be required for articles to be excluded. The design screening will determine whether articles assess the influence of proximity to a source using one or more comparison groups (in the case of categorical exposure data) or a range of distances (in the case of continuous exposure data). Where a system features unidirectional flow of material (e.g. a river system), the direction from the source may be used as an alternate exposure variable or may be combined with distance as the exposure variable. For example, the prevalence or concentration of antibiotic resistance markers downstream of a wastewater effluent could be compared to the same measure upstream of that suspected source. In addition, it will be determined if the full text article is written in English. Articles will be screened at this level using the following questions:
-
(1) Is the article written in English?
-
(2) Does the study report proximity to or direction from a potential source of resistance elements?
-
(3) Does the study have a comparison group (i.e. samples taken distant from/upstream of the source) or compare across a range of distances (i.e. samples taken at a range of distances)?
Any English-language article for which the answers to questions 2 and/or 3 are ‘No’ will be excluded from further consideration. For non-English articles, we will determine if questions 2 and 3 can be answered on the basis of the abstract. If so, an effort will be made to translate into English those articles meeting the inclusion criteria, as resources allow. Where the answers to questions 2 and/or 3 are not clear on the basis of the abstract for non-English articles, we will judge whether translation of the full-text article is necessary and practical on a case-by-case basis. Any potentially relevant non-English articles which cannot be translated by our team or collaborators will be identified in the review results as such. All articles meeting the inclusion criteria at this screening level will be deemed relevant to the review question, and the quality of evidence provided will be assessed.
Example of a study that would be included in the review
An observational cross-sectional study that would meet the study inclusion criteria examined the concentration of genes conferring resistance to the pharmaceutical, tetracycline, in groundwater adjacent to a swine manure lagoon (Hong et al., Reference Hong, Yannarell, Dai, Ekizoglu and Mackie2013). The study quantified the copy number of the ARGs, tetQ and tetZ, in groundwater samples from three locations down-gradient from the manure lagoon compared with two locations up-gradient from the manure lagoon.
Example of a study that would not be included in the review
An observational cross-sectional study was carried out in Nicaragua, which examined the resistance to multiple pharmaceutical antibiotics in Escherichia coli isolated from effluent of two municipal wastewater treatment plants and well-water in the community (Amaya et al., Reference Amaya, Reyes, Paniagua, Calderón, Rashid, Colque, Kühn, Möllby, Weintraub and Nord2012). Because this study did not report the proximity or location of wells with relationship to the effluent sources, this study would be excluded from this review.
Data extraction
Data from each included study will be extracted and used to assess the overall evidence for the relationship between proximity to or direction from point sources and antibiotic resistance. Specifically, details on the type of sample, source and outcome will be recorded, including the environmental media or biological sample type tested, quantity of the sample, the agricultural or municipal wastewater source type, the bacterial species and/or genes analyzed, and the microbiological method used. We will extract the sample size for each group, if exposure is categorical (e.g. near/distal or upstream/downstream), or total sample size, if exposure is continuous (e.g. km from source). If a statistical model was used to draw inference for the relationship of interest, then we will record the model type, effect measure, measure of variability and P-value. Use of methods to control for confounding or account for clustering in the data will also be recorded. If no effect measure was reported, but sufficient raw data are available, raw data will be extracted and, if possible, an appropriate effect measure computed. Due to expected heterogeneity in data types and study designs, the form of raw data to be extracted and the analytic method to be used will be decided on a case-by-case basis by at least two reviewers.
Whether a proportional measure or an absolute measure of resistance was used will be recorded. If the measure is proportional, it will be noted if the denominator was presented and, if so, whether the denominator was stable across sampling sites. Because it is not known whether the proportional measure alone, the absolute measure alone or the two in combination are most meaningful with regard to the human, animal and ecological risks posed by environmental resistance, this research decision will not be considered in the risk of bias assessment and all types will be included in review results as evidence pertaining to this review question.
Risk of bias assessment
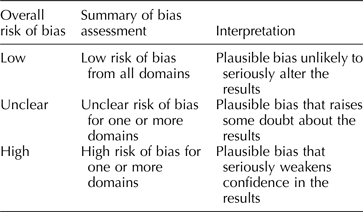
Each included study will be assessed for threats to internal validity. Risk of bias for each study will be assessed in three domains: sample selection bias, information bias, and confounding. For each domain, two team members will independently judge each study as having either a low, high, or unclear risk of bias, agreement required. For each study, overall risk of bias will be assessed by combining risk of bias from each domain (Table 3). This format for assessing risk of bias is adapted from the Cochrane Collaboration Risk of Bias Tool (Higgins & Green, Reference Higgins and Green2011) created for quality assessment of randomized controlled trials in human subjects. Consideration of domains of bias (i.e. confounding, selection bias), as opposed to specific methods of bias control (i.e. randomization), has been recommended as a flexible and valid way to apply established systematic review methodology to questions of veterinary and agricultural interest (Sargeant and O'Connor, Reference Sargeant and O'Connor2014b). This flexibility will permit us to use a single risk of bias tool for experimental and observational studies.
Table 3. Rubric for determining overall risk of bias for an included study, adapted from the Cochrane Collaboration Risk of Bias Tool (Higgins and Green, Reference Higgins and Green2011)
Sample selection bias
Threats to internal study validity due to sample selection bias, also known a survey bias, will be assessed using the following question: Were sample locations and sampling methods implemented such that sampling did not introduce systematic differences between the comparison groups (in the case of categorical exposure measures) or depending on the value of the exposure variable for each sample (in the case of continuous exposure data)?
Criteria for the judgment of ‘Yes’ (i.e. low risk of bias) include:
-
• Method for determining the sampling locations is identical independent of exposure status (i.e. distance or direction from source);
-
• Restriction of sampling locations is applied in the same way regardless of exposure status (e.g. sampling sites are all agricultural fields with a similar type and level of historical use);
-
• Time between sampling at all sites is sufficiently close so as to render the outcomes measured at these sites comparable for the sample type in question.
Criteria for the judgment of ‘No’ (i.e. high risk of bias) include:
-
• Sampling locations for samples taken near or down-gradient from the source are selected differently than the those taken distant or up-gradient from the source (e.g. up-stream sample site is a mid-channel location accessed by a boat while down-stream samples are collected via an existing monitoring station located at the river bank);
-
• Restriction of sample locations is applied differently depending on exposure status (e.g. agricultural fields selected for the unexposed comparison group, more distant from source, are restricted to only those with no previous history of manure application while fields selected for the exposed group, closer to the source, are not restricted to a particular manure use history.
Risk of bias from sample selection will be designated as ‘Unclear’ (i.e. unclear risk of bias) if there is insufficient information to permit judgment of ‘Yes’ or ‘No.’ If the method for selecting sampling sites or site characteristics are not described in sufficient detail, then it may not be possible to determine the risk of bias.
Information bias
Risk of bias due to systematic differences in the means of ascertaining the resistance outcomes between comparison groups, known as information bias or differential laboratory error, will be assessed using the following question: Were outcome ascertainment methods (i.e. methods of gene or bacterial measurement) conducted in a way that ensures the same accuracy regardless of distance or direction from the source(s)?
Criteria for the judgment of ‘Yes’ (i.e. low risk of bias) include:
-
• Identical microbiological methods applied to all samples;
-
• If results vary by laboratory factors (e.g. which laboratory, technician, testing date, or instrument used), a means of balancing laboratory factor between comparison groups was employed;
-
• Blinding of laboratory staff to exposure status.
Criteria for the judgment of ‘No’ (i.e. high risk of bias) include:
-
• Application of different microbiological methods depending on comparison group;
-
• Variable laboratory factors were known to be different between comparison groups (e.g. microbiological methods for all upstream samples were performed in one laboratory and for all downstream samples in a different laboratory).
Risk of information bias will be designated as ‘Unclear’ (i.e. unclear risk of bias) if there is insufficient information to permit a judgment of ‘Yes’ or ‘No’ on whether factors related to outcome ascertainment may have resulted in substantial bias.
Confounding
Risk of bias in the study results due to potential confounding will be evaluated using the following question: Were adequate methods to control for potential confounding employed?
Criteria for the judgment of ‘Yes’ (i.e. low risk of bias) include:
-
• Randomization of exposure;
-
• Restriction of the sample population;
-
• Analytical confounding control (e.g. stratification, regression adjustment).
Criteria for the judgment of ‘No’ (i.e. high risk of bias) include:
-
• Where confounding is likely, lack of any confounding control methods described above;
-
• Inappropriate method of confounding control used to address the potential confounder;
-
• Confounding control for some potential confounders is adequately implemented, but other important confounders are not addressed.
Risk of confounding bias will be designated as ‘Unclear’ (i.e. unclear risk of bias) if there is insufficient information on likely sources of confounding to permit judgment of ‘Yes’ or ‘No.’ This may be the case if, for example, reference is made to a confounding control method, but it is not described in sufficient detail to determine whether the method was correctly implemented.
Examples of potential confounders for the measurement of ARGs and ARBs in environmental samples are fluctuations in bacterial population size at different sampling locations, environmental media composition (soil type, water salinity), recent weather events differentially affecting sample sites, sample composition or history, landscape features, and other sources of antibiotics or antibiotic resistance factors.
Evidence synthesis
We will synthesize and present the overall state of the evidence on the review question narratively. For synthesis, evidence will be stratified into groups depending on which confounders were controlled for, outcome type and other relevant factors. If there are a sufficient number of high-quality studies with a low degree of heterogeneity, then meta-analysis of the data will also be conducted within strata. However given the broad nature of the research review question, we do not expect meta-analysis to be possible. Risk of bias across studies, due to publication bias for example, will be assessed within strata using funnel plots where possible.
Discussion
This article defines a protocol to evaluate the level of evidentiary support in the published, scientific literature for an effect of point sources of anthropogenic effluent on the prevalence or concentration of bacterial antibiotic resistance in the environment. This protocol details pre-specified criteria by which available evidence on the review question will be gathered and evaluated and is provided here for the purposes of transparency and completeness of this systematic review process. This protocol also serves as an example of how this rigorous knowledge synthesis method can be applied to a question pertaining to the ecology of antibiotic resistance in the environment.
Non-interventional environmental science research of this type shares important characteristics with other fields that must rely on observational (i.e. non-experimental) research to provide evidence about relationships of interest. In general, observational research is subject to bias in the estimation of etiologic relationships from a greater range of potential sources than randomized studies. For this reason, we have used an approach that assesses the risk of bias to the estimated relationship between point sources and increases in environmental ARB or ARG that is focused on the major domains from which such bias might arise, namely selection bias, information bias and confounding. These domains have been identified as the most important sources of bias in conservation and environmental management research (Pullin and Stewart, Reference Pullin and Stewart2006). Consideration of domains of bias, as opposed to specific methods of bias control (e.g. randomization), has been recommended as a flexible and valid way to apply established systematic review methodology to questions of veterinary and agricultural interest (Sargeant and O'Connor, Reference Sargeant and O'Connor2014a).
Originally developed in the human clinical context, systematic review and meta-analysis methodology has been increasingly applied to diverse fields, such as veterinary medicine (Sargeant and O'Connor, Reference Sargeant and O'Connor2014b), environmental management (Cook, Possingham and Fuller, Reference Cook, Possingham and Fuller2013), education (Best et al., Reference Best, Knight, Lietz, Lockwood, Nugroho and Tobin2013), toxicology (Birnbaum et al., Reference Birnbaum, Thayer, Bucher and Wolfe2013), and infectious disease ecology (Irwin et al., Reference Irwin, Yoon, Wang, Hoff, Zimmerman, Denagamage and O'Connor2011). To our knowledge, this protocol represents the first application of current systematic review methodology to antibiotic resistance in environmental systems. Rigorous knowledge synthesis in this important and rapidly expanding field, both on the proposed question and others, will be valuable to summarize the state of knowledge and to inform the design and focus of future studies.
Acknowledgments
The authors would like to thank Annette M. O'Connor for her helpful comments during the development of this protocol. This work is funded by the National Pork Board (Agreement no. 13-260) who requested a systematic review of the topic. The funding organization has been advised of the study question, but will play no role designating eligibility criteria, evaluating evidence or interpreting findings.







