Introduction
Salmonellosis is a major cause of human food-borne outbreaks and the second most commonly reported gastrointestinal infection in the European Union (EU) and European Economic Area (EEA) [1]. Among the Salmonella enterica subspecies enterica, S. Give is a rare serotype in the EU. Between 2010 and 2015, 684 cases of S. Give were reported by 18 EU and EEA countries [2]. The annual number of confirmed S. Give cases during this period ranged from 98 to 146, with Italy accounting for the highest number of cases (28%), followed by the UK (17%) and France (16%). Between 2007 and 2015, an average of 0.78 cases was reported annually in Malta.
Human outbreaks of S. Give have previously been documented in France in 2008 linked to formula milk [Reference Jourdan3] and in Germany in 2004 linked to the consumption of raw pork mince [Reference Jansen4]. S. Give has previously been isolated in chilli powder [Reference Wang5], beef carcasses [Reference Perez-Montano6], livestock [Reference Roy7, Reference Higgins8] and camels [Reference Raufu9].
The event
Between 17 and 22 October 2016, three sporadic cases of S. Give were notified to the Infectious Disease Prevention and Control Unit (IDCU) and this was followed by the confirmation of a cluster of five cases linked to a local restaurant on 26 October. An outbreak control team was set up to identify the source and implement control measures. The outbreak control team consisted of epidemiologists, environmental health inspectors, veterinarians and microbiologists.
Methods
Epidemiological investigations
We conducted a descriptive epidemiological investigation. Confirmed cases were defined as persons with laboratory-confirmed S. Give infection with disease onset since 1 October 2016. Suspected cases were defined as persons with gastroenteritis with disease onset since 1 October 2016 and epidemiologically linked to a confirmed case. To enhance case-finding, physicians working in the Accident & Emergency Department (A&E) at the main public hospital in Malta were requested to report all patients presenting with fever and gastroenteritis since 1 October 2016. Local laboratories were alerted to report all preliminary Salmonella results directly to the IDCU during the same period. We collected data on risk exposures from cases by telephone interviews using questionnaires.
Environmental investigations
Once a restaurant, snack bar or other food outlet was linked to a suspected or confirmed case, the IDCU informed the Environmental Health Directorate (EHD) who inspected the premises and collected environmental and food samples. Food trace-back investigations were conducted. These investigations included an assessment of all suppliers and inventory products used by implicated restaurants to identify common food items or food products.
Veterinary investigations
The National Veterinary Laboratory (NVL) was contacted to establish whether any S. Give isolates had been detected during routine sampling in 2016 based on the Salmonella National Control Programme in Malta as per the Commission Regulation (EU) 200/2012 [10]. As part of the EU control programme for Salmonella, all commercial registered broiler flocks are tested at 3 weeks of age (approximately 2 weeks prior to slaughter), according to the regulation 2160/2003 [11].
Microbiological investigations
Specimen collection
Suspected human cases were asked to submit stool samples for bacterial culture and antimicrobial sensitivity testing as part of the outbreak investigation. The EHD collected food samples, swabs from equipment/utensils and hand swabs from food handlers at the implicated restaurants.
At the local food manufacturer, food samples and swabs from equipment/utensils, hand swabs and stool samples from food handlers were collected by the EHD during onsite inspections at the food manufacturer's premises.
In 2016, as part of routine sampling for Salmonella, the NVL collected boot swab and dust samples from local broiler and layer farms across Malta.
Identification and genomic analysis
All human specimens were analysed in accordance with ISO ISO6579:2017 or following a locally adapted and validated protocol from the Public Health England (PHE) standards for microbiology investigations. After the identification of Salmonella infection by culturing the samples, isolates were serotyped based on the immunoreactivity of two cell surface structures, the O and H antigens and the S. Give serotype was assigned following the Modified Kauffmann–White Scheme [Reference Grimont and Weill12].
Following identification, a representative number of human, environmental and animal samples were selected for sequencing based on funding availability and those isolates most likely to support our working hypothesis based on epidemiological and environmental investigations. S. Give genomes collected from human cases were prepared in accordance with the Illumina TruSeq Nano DNA Sample Preparation Guide for Illumina Paired-End Multiplexed Sequencing [13] loaded onto an Illumina NextSeq Mid Output Flow Cell and sequenced using a 150 bp Paired End.
Selected food, environmental and animal isolates were fully sequenced. Strains were grown in 10 ml Brain Heart Infusion broth at 37 °C for 24 h. Cell pellets were obtained by centrifugation. The pellets were then washed and dissolved in 200 µl DNA/RNA Shield (Zymo Research, Irvine, California, USA). DNA isolation was outsourced and the isolates were commercially sequenced on a HiSeq 2500 sequencer (BaseClear, Leiden, The Netherlands).
The paired FASTQ files were processed using the standard bioinformatics pipeline used by the Gastrointestinal Bacteria Reference Unit (GBRU) of PHE for Salmonella isolates [Reference Ashton14]; (i) reads were trimmed and quality assessed using Trimmomatic [Reference Bolger, Lohse and Usadel15]; (ii) the species and subspecies identification of isolates were confirmed using the KmerID package [16]; (iii) sequence type (ST) and MLST profile were identified using the whole-genome sequencing (WGS)-mapping approach (MOST) package [Reference Tewolde17]. Antimicrobial resistance (AMR) profiles were predicted using the GeneFinder package (Doumith, unpublished); (iv) based on the ST of the isolates, they were associated with eBurstGroup (EBG) 67, i.e. Salmonella Give [Reference Achtman18] and mapped against the reference genome ERR1046255 downloaded from the National Center for Biotechnology Information's Short Read Archive [19] using Burrows–Wheeler Alignment [Reference Li and Durbin20]. Single-nucleotide polymorphism (SNP) variants of high quality were identified using the Genome Analysis Toolkit (GATK) Unified Genotyper [Reference McKenna21]. SNP distance and single-linkage clustering to sequenced S. Give isolates previously identified by PHE were determined using the SnapperDB [Reference Ashton22]. SNP alignment for the outbreak and PHE strains was generated from SnapperDB and used for generating maximum-likelihood trees using RAxML [Reference Stamatakis23]. The SNP variation within the outbreak cases, food and veterinary samples was analysed and a cluster was defined as sequences which presented within a 10 SNP difference.
Results
Epidemiological investigations
Between 1 October and 15 November 2016, 36 cases (21 confirmed and 15 suspected) of S. Give were detected in Malta (Fig. 1). Median age of cases was 45 years (range 15–88); 55% of cases were female.
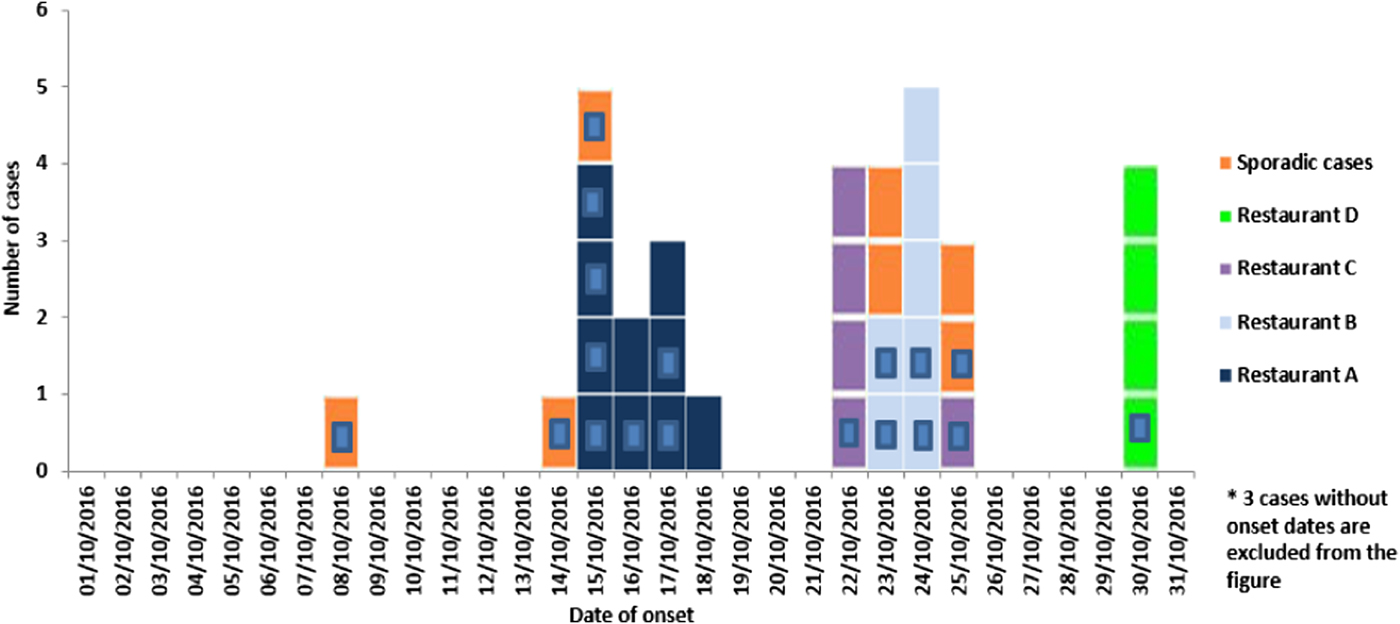
Fig. 1. Number of suspected and confirmed (blue square inside) Salmonella Give cases by restaurant exposure reported in October 2016 in Malta by onset of disease (n = 36*).
Symptoms of the patients included diarrhoea (93%), abdominal cramps (68%), fever (64%), vomiting (57%), nausea (39%) and headaches (36%) with a mean symptom duration of 5.3 days (range 2–12). Symptoms were generally severe with 10 (48%) of the confirmed cases requiring hospitalisation. One case developed septicaemia. Median age of hospitalised cases was 52.5 years (range 24–68). Twenty-six (72%) cases (12 confirmed and 14 suspected) were linked to four different restaurants (A, B, C and D) across Malta. All 26 cases reported eating ready-to-eat antipasti (bean dip and/or stuffed olives) from one of the implicated restaurants. The highest number of cases was reported in restaurant A (n = 10) followed by B (n = 7), C (n = 5) and D (n = 4). Seven (19%) confirmed cases were sporadic and not epidemiologically linked to other cases, food items or restaurants. Two British cases with a travel history to Malta during October 2016 were linked to the outbreak through WGS and met the case definition for a confirmed case. Thus, both cases are included in the overall confirmed case count.
Environmental investigations
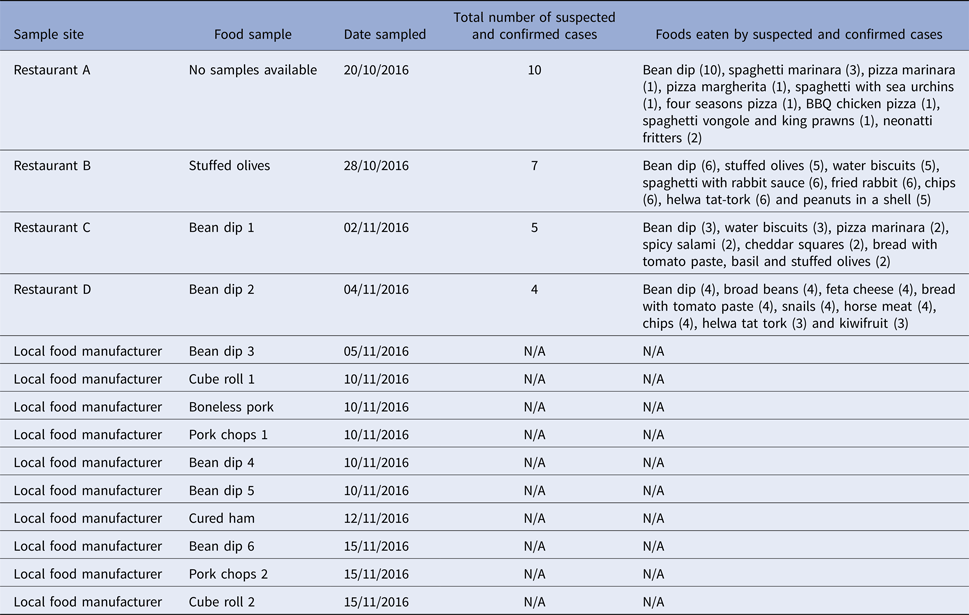
All four implicated restaurants (A, B, C and D) were supplied with bean dip and stuffed olives products deriving from the same local food manufacturer. From 28 October to 15 November 2016, 13 food isolates were positive for S. Give; three samples from restaurants B, C and D and 10 samples from the local food manufacturer (Table 1). No positive food samples were detected at restaurant A as leftover food samples were not available for testing. S. Give was isolated from stuffed olives stored in open containers in restaurant B and in bean dips from restaurants C and D. The stuffed olives and bean dips were all supplied by the same local food manufacturer. Following these findings, food-trace-back investigations were initiated and S. Give was first isolated from one batch of bean dip (bean dip 3 and 4) at the local food manufacturer on 5 and 10 November 2016, respectively. Sampling of additional food items at the local manufacturer isolated S. Give in a second batch of bean (bean dip 5 and 6) and raw meat products (Table 1).
Table 1. Salmonella Give-positive food samples by sample site, date sampled, number of suspected and confirmed cases and food exposures
Onsite inspection at the local food manufacturer
EHD identified a number of deficiencies in food safety management systems (FSMS) during the manufacturing of a ready-to-eat bean dip product: (i) inadequate separation of manufacturing areas between ready-to-eat and raw meat foods; (ii) equipment and utensils used for the preparation of raw meat was found in the ready-to-eat area; (iii) specific containers used for mixing bean dip ingredients were used and stored in the butchery area where raw meat was handled. Following these findings, the EHD in collaboration with the NVL, conducted an internal audit of the food manufacturer's premises and requested a full review of their internal FSMS procedures.
Product recalls
On 11 November 2016, a recall of the two incriminated batches of bean dip was implemented. All bean dip production was temporarily suspended until the Hazard Analysis Critical Control Point (HACCP) had been reviewed. This bean dip product was exclusively supplied to catering establishments only and not to retail outlets. As part of trace-back investigations for the cured ham product, the EHD inspected the premises of the main local meat supplier of the food manufacturer and collected environmental and food swabs. On 16 November, the EHD seized and tested a number of sealed food items at the local manufacturer including batches of cured ham and other meat products, provided by various suppliers. Batches of the cured ham product were not distributed to retail outlets.
Food handler stool and hand swab samples
A stool sample was collected from all five food handlers working at the local food manufacturer. Of these, one (25%) tested positive for S. Give despite the food handler reporting no history of gastroenteritis illness in the previous few weeks. This food handler was excluded from returning to work until three consecutive stool cultures collected not less than 24 h apart were negative for Salmonella [Reference Heymann24]. All five food handlers who provided stools possessed the relevant food hygiene safety certificates upon inspection. In addition, no medical absentees from gastrointestinal illness were reported among any of the five food handlers during the outbreak period. Of four swabs taken from food handlers, one (25%) glove swab taken from a food handler during raw meat handling, tested positive for S. Give.
All known links established during the epidemiological and environmental investigations are summarised in the following network (Fig. 2):
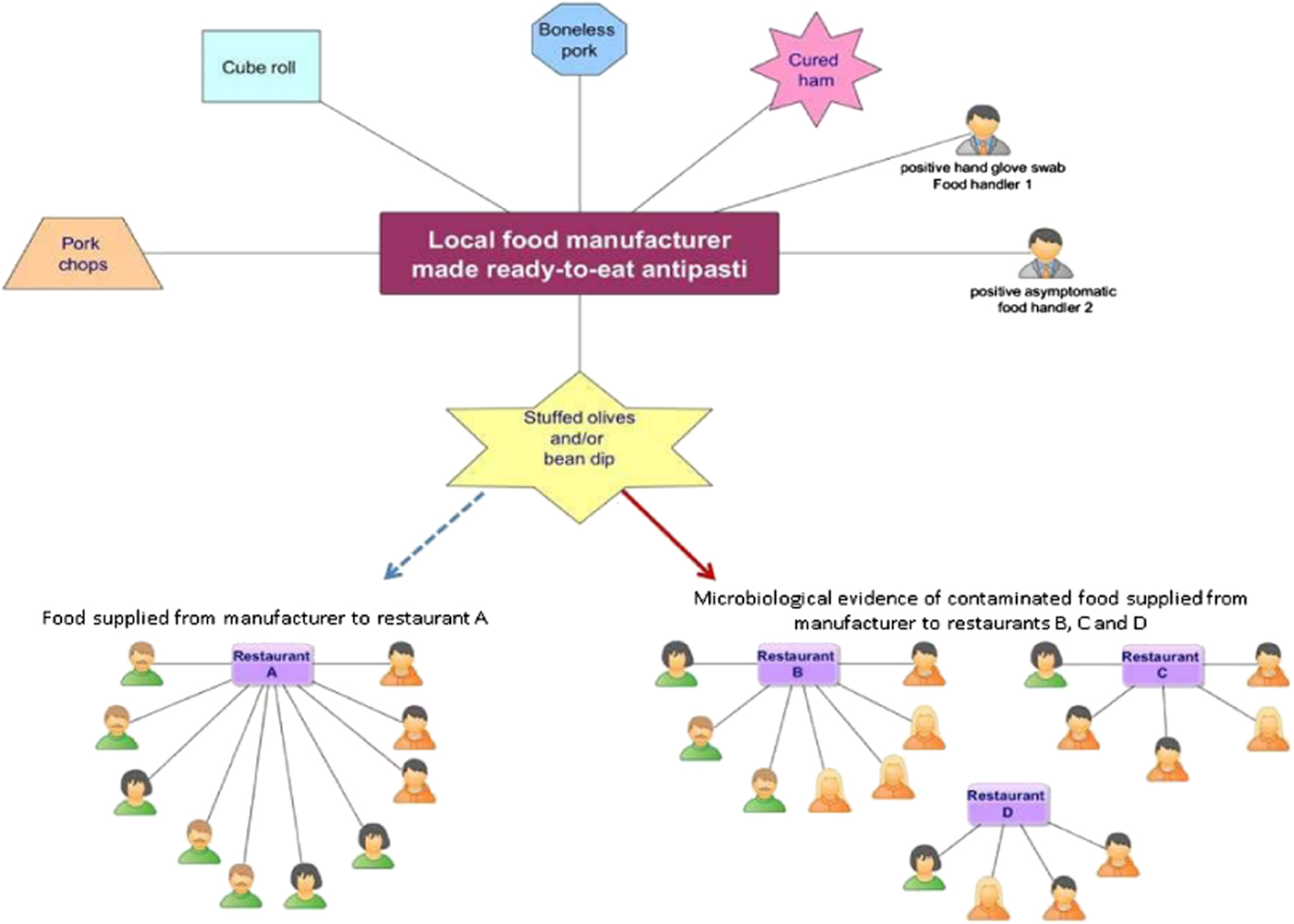
Fig. 2. Network diagram showing all known epidemiological links between the 26 confirmed (green) and suspected (orange) human cases by gender, four restaurants (A, B, C and D) and positive environmental/food samples originating from the local food manufacturer.
Veterinary investigations
Between 1 April and 13 October 2016, the NVL detected eight positive S. Give isolates (six from chicken broilers and two from layer flocks) from four farms across Malta. Six of the positive isolates were detected from 29 September to 13 October. In addition, on 14 October 2016, S. Give was isolated from a swine carcass during random swabbing at a slaughter house. The swine originated from a local farm that is one of the suppliers of pork to a meat processing company that supplies the local food manufacturer under investigation. No positive S. Give samples were detected in batches of pork products sampled at the meat processing company.
Following the detection of positive S. Give isolates among chicken broilers and layer flocks, the Veterinary Regulation Directorate (VRD) implemented over 60 corrective measures as part of their annual checks on bio-security measures. These corrective measures included ensuring appropriate housing structures, netting, pest control, disinfection pits and boot washes in front of houses.
Microbiological investigations
Specimen collection results
From 1 October to 15 November 2016, 18 human specimens were collected from suspected human cases. Between 20 October and 4 November 2016, the EHD collected 23 food samples, 19 swabs (12 from equipment/utensils and seven hand swabs from food handlers) from restaurants A, B, C and D, respectively. At the local food manufacturer, 45 food samples and 18 swabs (14 equipment/utensils and four hand swabs from food handlers) were collected by the EHD during three separate visits to the manufacturer's premises on 5, 10 and 15 November 2016, respectively. In addition, on 15 November, five stool samples were collected from food handlers.
In total, of 97 specimens collected during the epidemiological and environmental investigations in Malta, 33 (34%) were positive for S. Give. Of these, 19 (58%) were human, 13 (39%) food and one (3%) was an environmental hand swab. During veterinary investigations, of 502 specimens collected from broiler and layer flocks by the NVL during routine sampling in 2016, eight (1.6%) were positive for S. Give.
Identification and genomic analysis results
Of 44 positive S. Give isolates identified during the epidemiological, environmental and veterinary investigations, 28 (64%) were selected for the MLST analysis. Two sequences could not be validated and did not pass the quality test for inclusion in the MLST analysis. In total, 13 human-confirmed outbreak cases (sample collection dates; 13 October–15 November 2016) and one non-outbreak case (retrieved from the PHE database; a British person with travel history to Spain), four food samples and one environmental swab (sample collection dates: 1–15 October 2016) and seven veterinary isolates (sample collection dates: 29 September–13 October 2016) were included.
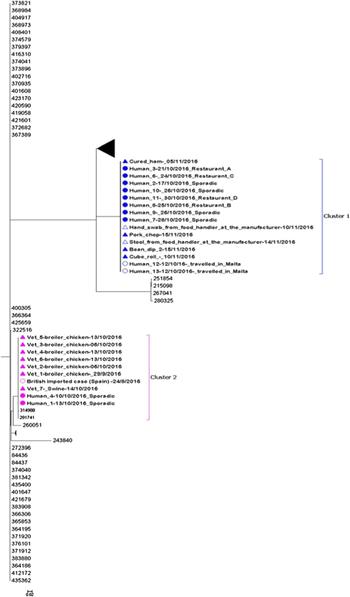
The whole-genome MLST sequencing analysis of the remaining 26 sequences indicated that there were two distinguishable clusters. SNP analysis confirmed the close relationship within both clusters observed in the maximum likelihood tree (Fig. 3). The two clusters were distinguishable from each other with an average of 6374 SNPs difference.
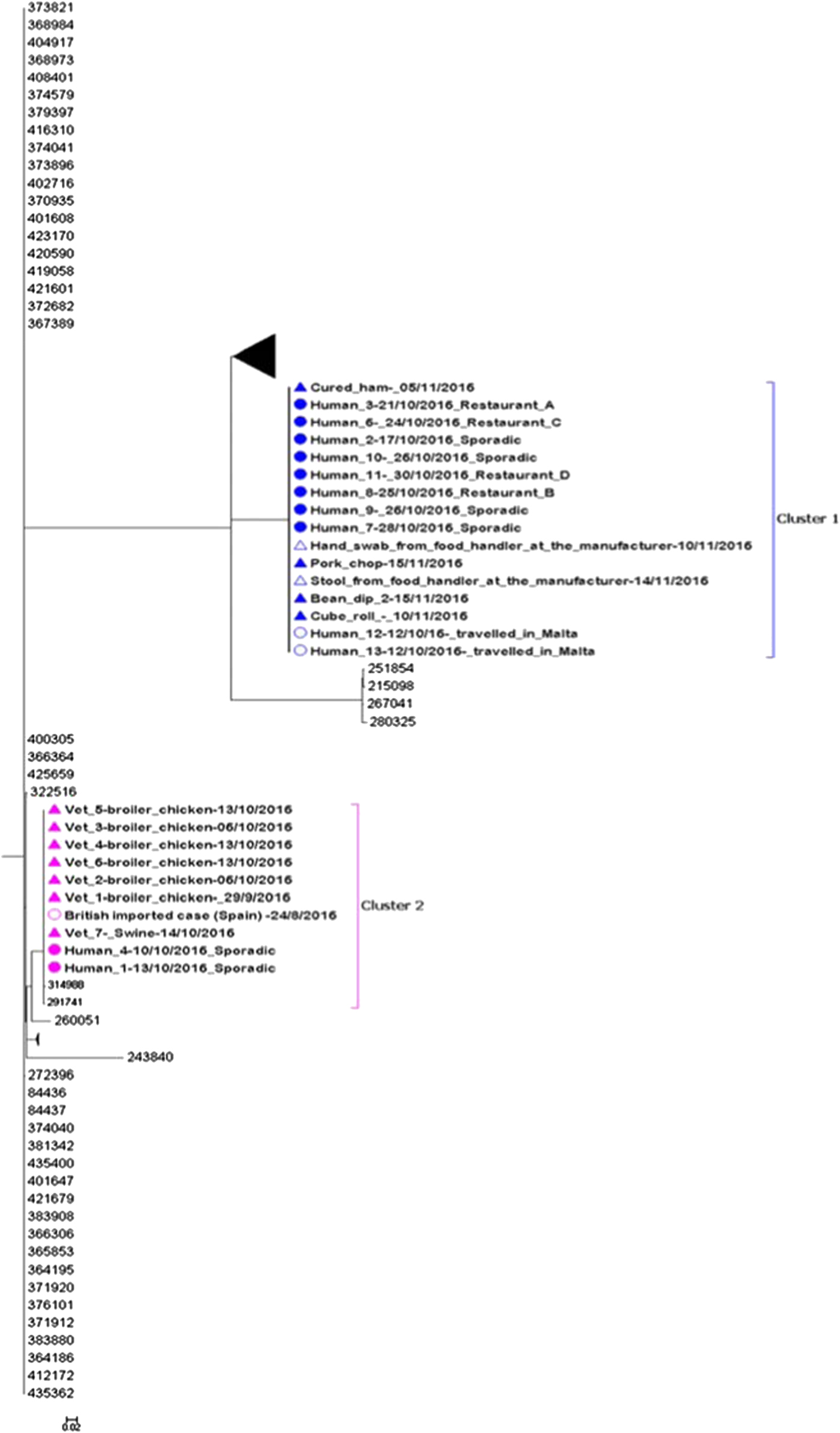
Fig. 3. Maximum likelihood tree of complete sequences collected from confirmed case isolates (UK and Malta), food and environmental isolates originating from the local manufacturer and veterinary isolates (broiler chickens and swine). Each isolate in the tree is labelled with the date of sample collection. Human isolates in clusters 1 and 2 are labelled with restaurant exposure or as sporadic if known. The bootstrap values are shown.
Eleven of 13 sequenced human isolates clustered together with four food isolates and one glove swab from a food handler originating from the local food manufacturer (Fig. 3: cluster 1). The SNP difference range within this cluster was 0–2 SNPs. Ten of the sequenced human isolates from Maltese confirmed cases from cluster 1 were epidemiologically linked to restaurants A, B, C and D. No epidemiological link could be established between the two exported British cases and restaurants A, B, C and D.
Two sequenced human isolates from Malta and one from the UK (a British person with travel history to Spain in August 2016) clustered together with seven veterinary isolates (Fig. 3: cluster 2). The three human isolates and the swine isolate were all indistinguishable and the SNP difference range within this cluster was 0–7 SNPs. No epidemiological links could be established between the three human and eight veterinary isolates in cluster 2.
AMR analysis
All human isolates included in the MLST analysis in clusters 1 and 2 were resistant to monoglucoside.
Discussion
We describe the first documented national outbreak of S. Give in Malta. Despite the initial hypothesis of a unique outbreak caused by the same strain, WGS investigations indicated that two genetically separate S. Give strains were present among the confirmed cases with two distinct clusters identified. Genetic analysis linked cluster 1 to contaminated food in restaurants that were all supplied by the same local food manufacturer. Cluster 2 was genetically associated with infected animals from local farms but no further epidemiological links could be identified.
In cluster 1, a range of 0–2 SNPs differences were identified linking eight human cases, the bean dip product and the other environmental samples collected from the same local food manufacturer. In addition, the same S. Give strain was identified in the stool from an asymptomatic food handler and from a glove swab taken from another food handler during raw meat handling, strongly suggesting that cross-contamination at the local food manufacturer was the likely source of infection for cluster 1. This hypothesis was also supported by environmental investigations which identified a number of deficiencies during the manufacture of the ready-to-eat bean dip product. Measures implemented included conducting an internal audit of the local food manufacturer's premises including a full review of their internal FSMS procedures, temporary suspension of all bean dip production until the HACCP had been reviewed and a product recall of the implicated bean dip batches.
Previous outbreaks of gastrointestinal illness following cross-contamination of raw and ready-to-eat foods during the manufacturing process have been documented [Reference Okpo25, Reference Matsui26]. This suggests that adherence to food safety practices at manufacturing level are essential in order to limit food contamination.
Despite the seemingly localised nature of this outbreak, WGS analysis indicated that two British citizens who travelled in Malta during October 2016 were also part of cluster 1, suggesting they may have acquired the infection in one of the implicated restaurants. However, S. Give was only diagnosed once they returned to the UK and it was not possible to collect any additional information on possible risk exposures or establish an epidemiological link with any of the implicated restaurants.
Despite the initial hypothesis of a single unique outbreak, a genetically distinguished S. Give strain with a difference of more than 250 SNPs from the strain identified in cluster 1 was identified for cluster 2. In cluster 2, two human cases clustered together with veterinary samples collected from local farms with a range of 0–7 SNPs difference. This unexpected link was identified by including routinely collected veterinary samples in the WGS, underlining the importance of maintaining animal surveillance for Salmonella and highlighting the need for continued cross-collaboration between human, environmental and animal health sectors in Malta. In addition, a British case with travel history to Spain was genetically linked with the Maltese cases in cluster 2, suggesting a possible link. However, unfortunately, no additional travel history information could be obtained for this case, so it was not possible to establish any plausible link with the other human or animal cases.
The severity of symptoms and high hospitalization rates of patients in this outbreak indicates a high virulence of this specific serotype. A similar finding was found in previous human S. Give outbreaks [Reference Jourdan3, Reference Jansen4]. This outbreak also highlights a potential role for asymptomatic food handlers in the transmission of Salmonella infection and underlines the importance of testing asymptomatic food handlers during outbreaks of gastrointestinal illness. Previous Salmonella outbreaks involving asymptomatic food handlers as a potential source of infection have been well documented [Reference Holman27–Reference Dryden29].
WGS data are a useful tool for confirming the source of infection in outbreaks when food and human isolates are available. Previous food-borne outbreaks of Salmonella using WGS data to help identify the source of infection have been described [Reference Wilson30–Reference Pärn32]. In this specific outbreak setting, WGS analysis was also used to distinguish two potential different sources of infection which would otherwise be indistinguishable using traditional assays. Collaboration with European partners was essential for performing WGS during this outbreak investigation as it is currently not possible to perform such analysis in Malta.
The main limitation to this investigation was the lack of capacity to conduct WGS in Malta that delayed the outbreak investigation and response.
Conclusions and recommendations
Although the source of infection was not identified, epidemiological and environmental evidence pointed towards cross-contamination of raw and ready-to-eat foods at the local food manufacturer as the likely source of infection for this S. Give outbreak in Malta. WGS provided additional evidence to support this hypothesis (cluster 1). Concurrently, a second human cluster (cluster 2) with possible infection from positive animals was identified. To prevent future cases and outbreaks, adherence to food safety practices at manufacturing level need to be reinforced [33]. The establishment of an EU reference laboratory network for Member States with limited or no capacity to conduct WGS for early outbreak detection and response should be considered.
Acknowledgements
The authors gratefully acknowledge the contribution of the Environmental Health Directorate (EHD) for their work during the environmental investigations, Public Health Laboratory and the Pathology Department at Mater Dei Hospital for their contribution with the microbiological investigations, the National Veterinary Laboratory (NVL) and Veterinary Regulation Directorate (VRD) for their support with the veterinary investigations. The authors would like to thank the European Centre for Disease Prevention and Control (ECDC), Public Health England (PHE) and the Netherlands National Institute for Public Health and the Environment (RIVM) for their support with the WGS analysis. The authors also thank Kostas Danis from the European Programme for Intervention Epidemiology Training (EPIET) for providing useful comments on the article.
Author contributions
Epidemiological investigations were conducted by Alastair Donachie, Maria-Louise Borg and Tanya Melillo. Alastair Donachie drafted the manuscript. Hassan Hartman carried out the bioinformatics and contributed towards phylogenetic analysis of human, food and veterinary isolates. Laura Bubba coordinated the WGS analysis, phylogenetic analysis and created the network maps. All authors contributed to editing and final approval of the manuscript.
Conflict of interest
None.