INTRODUCTION
With a morbidity of 2–10 cases per million, invasive listeriosis is a rare disease; however, it may be a serious health risk for immunocompromised individuals and the elderly because Listeria monocytogenes is widely spread and may enter the food chain despite standard precautions [Reference Gillespie1–Reference Hjaltested3]. The reported figures depend on the sensitivity of surveillance, with the highest rates registered in countries with statutory notification of Listeria infections [Reference Hedberg4, Reference Doorduyn5]. An integral part of the monitoring system is the microbiological survey, conducted by a central reference laboratory, using phenotypic (serology) and genotypic (PFGE, MLVA and MLST) methods [Reference Liu6, Reference Allerberger7]. Since most isolates belong to serovars 4b, 1/2b, and 1/2a, the discriminatory power of Listeria serology is relatively low. However, the genetic division of the serovars into lineages and phylogenetic groups has increased the usefulness of serotyping as a relatively quick, low-cost method for routine microbiological surveillance [Reference Doumith8].
In Israel, invasive listeriosis has been reported in immunocompromised patients and pregnant women [Reference Tweezer-Zaks9–Reference Siegman-Igra12]. Notification became mandatory in 1996, and the Reference Listeria Laboratory (RLL) was established in 1997. The aim of this report is to present the first longitudinal survey of clinical L. monocytogenes isolates, tested at the RLL.
MATERIALS AND METHODS
The study includes a total of 321 L. monocytogenes isolates from sporadic cases, identified by hospital laboratories, using standard methods [Reference Kerr and Lacey13] over a period of 11 years (1997–2007). Using the data in the accompanying forms, they were divided into perinatal and non-perinatal, and the second group was subdivided into strains from patients aged <60 years and ⩾60 years (Table 1). The isolates were from blood, CSF, maternofetal (amniotic fluid, placenta and products of conception) and focal sites (pleural and peritoneal fluid).
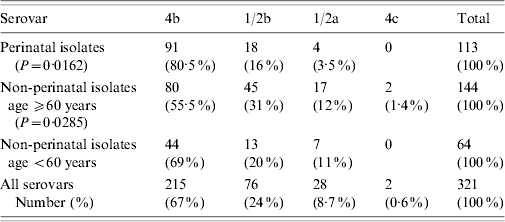
Table 1. Serovar distribution of L. monocytogenes isolates in the host groups
Biochemical verification was carried out with the Microgen Listeria-ID system (Microgen Bioproducts Ltd, Camberley, Surrey, UK). Serotyping was performed with commercial antisera, following the manufacturer's protocol (Denka Seiken, Tokyo, Japan), with a modification for the H-antigen agglutination test [Reference McLauchlin and Shah14].
Antimicrobial susceptibility was checked by the disk diffusion method in accordance with the recommendations of the Swedish Reference Group for Antibiotics [15]. Commercially prepared disks (Oxoid, Basingstoke, UK) were used, containing ampicillin (AMP, 10 μg), sulphamethoxazole–trimethoprim 19.1 (SXT, 25 μg), vancomycin (VA, 30 μg), tetracycline (TE, 30 μg), gentamicin (CN, 10 μg) and chloramphenicol (C, 30 μg). The results were interpreted using the criteria of the National Committee for Clinical Laboratory Standards for Listeria spp. and Staphylococcus spp., as well as previously published data [Reference Troxler16–Reference Hansen, Gerner-Smidt and Bruun18]. Reference strains of Escherichia coli (ATCC 25922) and Staphylococcus aureus (ATCC 25923) were tested in each run.
Data analysis
Correlations between the general prevalence of the serovars and their distributions in the host groups were analysed by χ2 test (two-tailed tests were used). The morbidity estimates (per million) were calculated using the population figures of the Israel Bureau of Statistics [Statistical abstracts (http://www.cbs.dov.il)]. The incidence estimates (per 100 000) were based on the number of pregnancies and individuals aged <60 and ⩾60 years.
RESULTS
Perinatal isolates
A total of 113 (35%) isolates were from perinatal cases: 29 newborns and 84 pregnant women (mean age 29 years, range 19–40). The most prevalent serovar was 4b (80·5%), followed by 1/2b and 1/2a (P=0·0162, Table 1). The isolates were from blood (77, 68%), maternofetal sites (29, 26%) and CSF (7, 6·2%).
Non-perinatal isolates
Of the 208 (65%) non-perinatal isolates, 144 were from patients aged ⩾60 years (mean age 79 years, range 60–91) and 64 from patients aged <60 years (mean age 42 years, range 2–59). The most prevalent serovar was 4b, followed by significantly high numbers of 1/2b and 1/2a strains in the ⩾60 years subgroup in comparison with the general serovar distribution (P=0·0285, Table 1). Two 4c strains were identified in blood and CSF from female patients aged 80 and 81 years, respectively. Most of the samples were from blood (134, 78%), followed by CSF (29, 17%) and focal sites (9, 5·2%); 105 patients were male and 103 were female (M/F ratio=1·01).
The seasonal distribution showed that 206 isolates (64·2%) were submitted between May and October (P=0·0001).
Antimicrobial susceptibility
A total of 115 L. monocytogenes strains were checked, of which 96 (83·5%) were from blood, 13 (11·3%) from CSF, and 6 (5·2%) from maternofetal sites. The serovar distribution was as follows: 4b (80, 69·5%), 1/2b (17, 15%), 1/2a (16, 14%) and 4c (2, 2%). Resistance to TE was found in eight 4b isolates (6·5%), of which seven were from blood and one from CSF. Three of the resistant strains were from perinatal cases and five from non-perinatal cases.
DISCUSSION
In our population 4b was the most common Listeria serovar isolated over an 11-year period. This result is consistent with its recognized high pathogenic potential [Reference Vazquez19]. As previously noted [Reference Vazquez19, Reference McLauchlin20], serovar 4b is more common in the perinatal group (P=0·0162), compared with those aged ⩾60 years (P=0·0285, Table 1). The different distribution of serovar 1/2b in the two groups may be connected to genetic variances in pathogenic tropism, since it is of the same lineage as 4b but in a separate phylogenetic group [Reference Doumith8].
The prevalence of 1/2a, which belongs to a different lineage (lineage II), differs from other reports where it has been the most common or the second most common serovar in clinical strains [Reference Gillespie1, Reference Goulet21]. Its occurrence may also be host dependent, in that it is less commonly isolated from perinatal cases, and increases in those aged ⩾60 years (Table 1).
Serovar 4c (lineage III) is rarely found in clinical isolates [Reference Roberts22, Reference Liu23]. In this study, both strains were rhamnose +, which places them in the genetically distinct and reportedly more virulent subgroup IIIA [Reference Roberts22]. However, low exposure to food contaminated with Listeria isolates belonging to the three subgroups of serovar 4c may be a reason for the small number of clinical cases described in our population [Reference Roberts22, Reference Ward24].
The noted summer increase in morbidity [Reference Gillespie1, Reference Siegman-Igra12, Reference Voetsch25] was also found in our study, with the May–October period being the hot season in Israel. According to one hypothesis, there is a link with the concomitant increase in gastrointestinal infections, which, in addition to provoking immunosupression, facilitate the entry and dissemination of L. monocytogenes in the intestine [Reference Crum2, Reference Schwartz26, Reference Roberts and Weidman27]. There is a well-documented case report of sporadic listeriosis in a healthy adult following Shigella sonnei shigellosis [Reference Lorber28], as well as experimental data of interactions between L. monocytogenes and other enteric bacteria at the level of the intestinal mucosa [Reference Jensen, Harty and Jones29]. The relevance of these findings to listeriosis incidence requires additional studies, considering the established high morbidity of shigellosis and other enteric diseases in Israel [Reference Vasilev30].
As in other reports, antimicrobial susceptibility is high, with tetracycline resistance most often described in human isolates when antimicrobial resistance is present [Reference Charpentier and Courvalin31]. Incidentally, a low incidence of tetracycline resistance also was found in food isolates in Israel from the same period (V. Vasilev, unpublished data).
Based on the number of submitted isolates, the morbidity of invasive listeriosis in Israel is 4·4 per million. Since 2005, the number of reported Listeria cases has been increasing (Fig. 1), while most industrialized countries report a decline [Reference Swaminathan and Getrner-Smidt32]. The increase in Listeria cases in Israel requires further study to understand its aetiology. As previously reported by others [Reference Gillespie1, Reference Hedberg4, Reference Vazquez19, Reference de Valk33], invasive listeriosis was more common in the perinatal age group (5·6/100 000) than in individuals aged ⩾60 years (1·5/100 000).
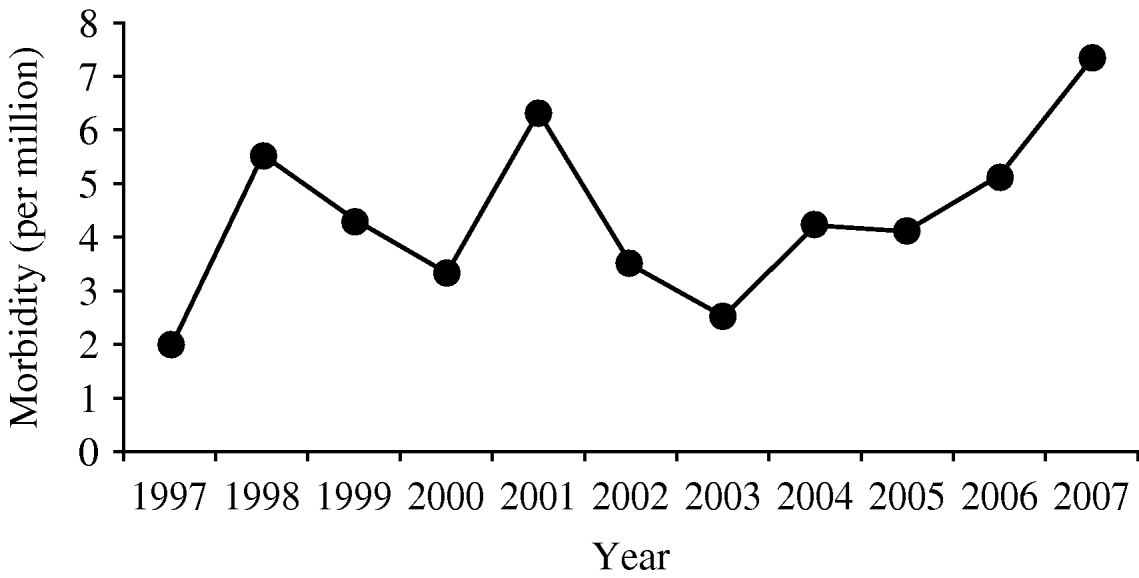
Fig. 1. Estimated annual morbidity (per one million).
ACKNOWLEDGEMENTS
We acknowledge the contribution of the late Dr A. Mates, former head of the Department of Laboratories of the Ministry of Health, who initiated the establishment of the RLL.
DECLARATION OF INTEREST
None.




