INTRODUCTION
Q fever is a common zoonotic infection caused by Coxiella burnetii, an obligate intracellular gamma proteobacterial organism [Reference Cutler, Bouzid and Cutler1]. Cattle, sheep and goats are the livestock most often implicated in human disease. Transmission to humans is believed to be particularly associated with contact with parturient animals. A majority of human cases of C. burnetii infection are asymptomatic [Reference Cutler, Bouzid and Cutler1]. Where symptomatic infection occurs, typical signs and associated symptoms are headache, pyrexia, and respiratory tract infection including atypical pneumonia. Hepatitis may also occur [Reference Parker, Barralet and Bell2]. Chronic infection is well recognized, usually in the form of Q fever endocarditis.
Although Q fever is present worldwide, the incidence of human disease appears to vary widely throughout the world. Compared to the rest of the British Isles, Northern Ireland reports particularly high rates of Q fever in humans. From 1997 to 2003, the number of laboratory reports of C. burnetii has ranged from 11 to 53 (0·7–3·1 cases per 100 000 population) which is several times higher than England and Wales [3].
The agric-food sector is considerably more important to the Northern Ireland economy than for other regions in the UK and grazing livestock farming is a key part of the Northern Ireland economy, forming 88% of the agricultural land use compared to 60% in the UK as a whole [4]. The cattle population comprises 1·65 million animals on 22 250 farms [5]. There are 286 000 dairy cows and 273 000 beef cows with most of the remainder of the cattle population being younger animals. The sheep population comprises of 2·02 million animals on 8646 farms [5].
The importance of cattle and sheep as the source of human infection may vary throughout the world. For example, data from England suggest that exposure to cattle, but not sheep, goats, cats, raw milk, or hay (all considered possible sources of Q fever) was associated with testing positive for C. burnetii IgG but this association was not independent of total farm animal contact [Reference Thomas6]. The authors of that study concluded that the risk of Q fever in livestock farms is related to contact with the farm environment rather than any specific animal exposure [Reference Thomas6]. In contrast, a literature review in the USA suggests that C. burnetii is enzootic in domestic ruminants and wild animals with widespread human exposure. Sheep and goats appear to be a more important risk for human infection in the USA than cattle or wild animals [Reference McQuiston and Childs7].
There is very little detailed epidemiological data regarding the distribution and determinants of Q fever infection in cattle from anywhere in the world. The organism is present in cattle populations from all regions of the world with the notable exception of New Zealand. In Italy, there has been a clear suggestion that the seroprevalence is higher in housed cattle than cattle kept outdoors [Reference Capuano, Landolfi and Monetti8].
The seroprevalence rates reported in cattle populations vary greatly, ranging from 3·4% to 84% [Reference McQuiston and Childs7, Reference Adesiyun, Jagun and Tekdek9–Reference Marrie13]. However, the literature is difficult to interpret because of the use of differing serological methods and assay cut-offs. For example in Japan, cattle seroprevalence of 1·1–3·9% was noted in the 1950s whereas in 1992 it was shown that 29·5% of healthy cattle and 84·3% of those with reproductive disorders were seropositive using immunofluorescence assay (IFA) [Reference Htwe14]. In the UK, using an ELISA for the detection of IgG to C. burnetii in bulk tank milk, 21% of 373 randomly selected samples from dairy herds in England and Wales, showed antibody evidence of C. burnetii infection [Reference Paiba15]. In Canada, 67% of dairy herds were seropositive using ELISA [Reference Lang16]. In USA, reported cattle seroprevalence varies from 1% to 73% [Reference Kim17]. Some seroepidemiological studies have recorded apparent increases in C. burnetii antibody seroprevalence in cattle over recent decades [Reference Lang and Marrie18]. However, in general the data on prevalence of C. burnetii infection in cattle is incomplete as there is insufficient surveillance [Reference McQuiston and Childs7].
Northern Ireland is particularly well placed to perform a study to examine the epidemiology of Q fever in cattle because of the comprehensive database on cattle via the Animal and Public Health Information System (APHIS) [Reference Houston19].
C. burnetii infection in cattle is generally perceived to have little agro-economic or animal health implications; however, infection may be associated with abortion [Reference Sting20, Reference Cabassi21]. There has also been a recent suggestion of a link to subclinical mastitis in dairy cattle [Reference Barlow22]. Some evidence of an effect on herd fertility has been suggested by studies on vaccines that have been developed for cattle showing that herd fertility may be increased when these are used [Reference Krauss23].
METHODS
Bovine serum specimens
Animals were selected from those undergoing statutory routine brucellosis testing (reproductive animals aged >1 year) using a two-stage process. Herds were selected from each of the ten Northern Ireland District Veterinary Office (DVO) regions within strata defined by herd size (small <50 animals, medium 50–100 animals, large >100 animals) and by herd type (dairy or beef). This selection process was based on all routine brucellosis herd tests submitted to the laboratory on certain dates with quotas (based on the DVO cattle population) for the criteria being predefined. All breeding animals in Northern Ireland herds aged >1 year are included in the Brucella testing protocol so the whole breeding cattle population is effectively sampled. Any samples that were reactive on the initial Brucella screening test, serum agglutination test (SAT) were unavailable to test (2–3% of the samples). The first 20 remaining blood samples in each selected herd were tested.
Bovine testing method
The assay used for bovine C. burnetii IgG testing was the Bommeli Chekit indirect ELISA (Bommeli, Switzerland). The standard manufacturer's protocol was followed for testing, use of controls and interpretation of results. All positive and equivocal samples were retested.
The Bommeli Chekit Q fever IgG kit was supplied with microtitre plates pre-coated with inactivated C. burnetii Nine Mile strain. Provided that optical densities (OD) for positive and negative controls fell within the manufacturer's stated guidelines the test was considered valid. An antibody index expressed as a percentage value was determined for each sample tested:
The results were then as follows: <30% negative, 30–40% equivocal, >40% positive.
Herds were considered to be positive if at least one animal tested positively. Results were examined by animal and by herd and the data were also analysed using a random intercept multilevel logistic regression model (MLwiN, version 1.1) which included two levels, herd and animal. This model allowed for a lack of independence of observations from cows within herds. The produced parameter estimates and standard errors (and consequently significance tests) were adjusted for any correlation of the observations from cows from within herds.
Characteristics at herd level that were analysed included herd type (beef or dairy), herd size, geographical area of farm holding (designated by the 10 DVO areas shown in Figure 1 and whether sheep were kept on the same farm holding (as determined by whether the farmer had a sheep herd number in addition to his cattle herd number). Characteristics at animal level that were analysed included breed, whether breed is a recognized dairy breed, sex, age at sampling (sampling date minus date of birth), length of time animal has been in the herd in which it was sampled and number of moves (herd-to-herd movements) recorded against the animal prior to sampling in the survey.
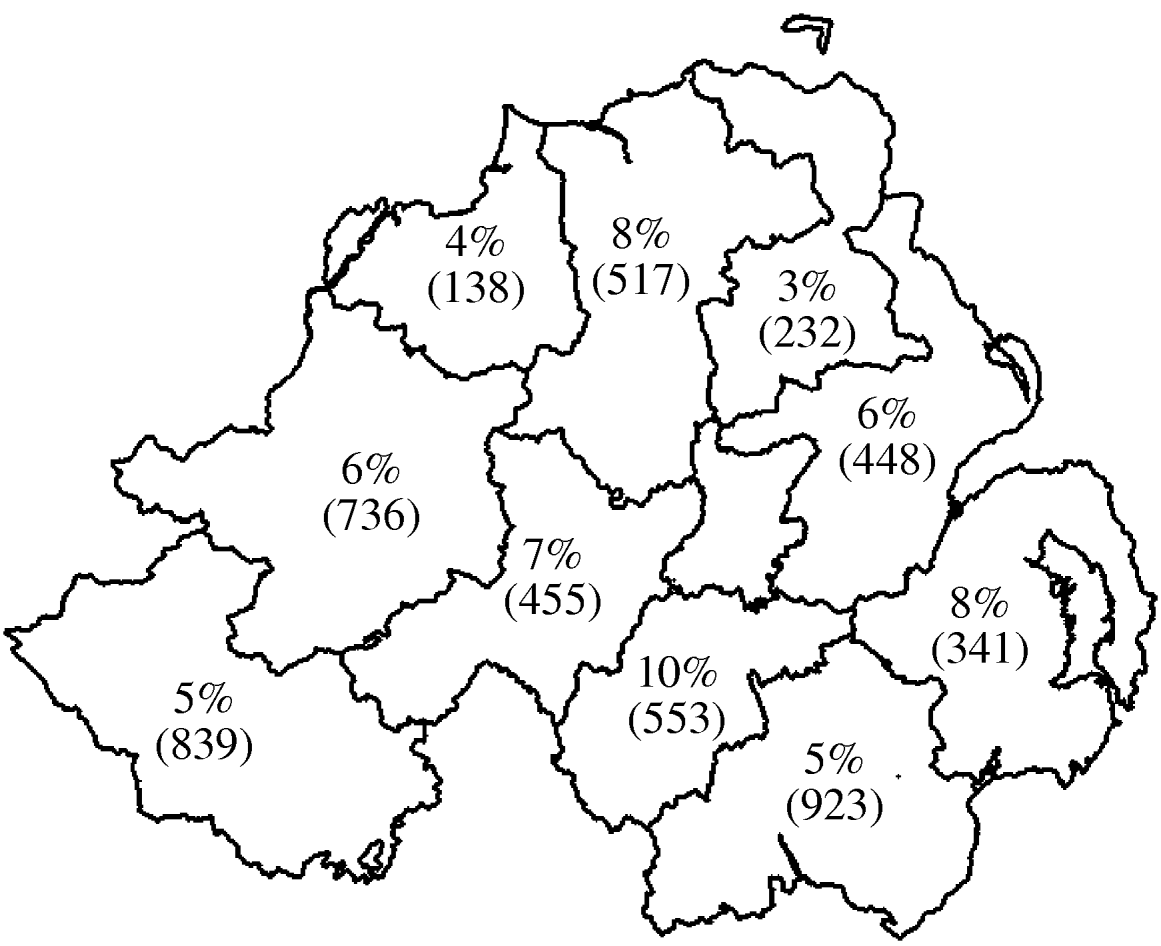
Fig. 1. Geographical distribution of seroprevalence at animal level. Values shown are percent of animals positive with number of animals tested in parentheses.
Multilevel modelling results are presented as the odds of animal seropositivity compared to reference categories. All factors (both animal and herd) are adjusted for each other in the model.
Validation of the Bommeli Chekit indirect ELISA
A panel of 83 selected bovine sera were used for validation of the ELISA and the results were compared with the bioMérieux indirect IFA (bioMérieux, France).
The bioMérieux IFA was performed according to the manufacturer's instructions. Briefly, sera were screened at 1/80. Relevant controls were included [the negative control was phosphate buffered saline (PBS), the positive control was a local high-titre antibody-positive bovine serum]. A volume of 10 μl of each dilution and relevant controls were added to the appropriate well on a C. burnetii-Spot IF slide (bioMérieux), incubated at 37°C for 30 min and then washed for 10 min in PBS. Conjugate was applied and the slide incubated for 30 min at 37°C, washed, mounted and examined. The conjugate used was Sigma rabbit anti-bovine IgG FITC labelled, used at a 1:300 dilution as recommended by the manufacturer (Sigma, USA). A positive reaction was one in which there was typical distinct fluorescence for >50% of C. burnetii present on the slide well under examination.
Testing of ovine, caprine, porcine, rat and mouse samples
Ovine (1022 samples), caprine (54 samples) and porcine (64 samples) sera were residual material from samples submitted from clinically normal populations, in the course of routine surveillance activities. The 31 rat (Rattus norvegicus) and 31 mouse (Mus musculus) sera were collected in County Down for previous work on hantavirus seroprevalence [Reference McCaughey24].
Ovine, caprine, porcine, rat and mouse samples were tested using C. burnetii-Spot IF slide (bioMérieux) indirect IFA using relevant conjugates (donkey anti-sheep IgG FITC used 1:80; rabbit anti-goat IgG FITC used 1:400; rabbit anti-pig IgG FITC used 1:32; goat anti-rat IgG FITC used 1:64; rabbit anti-mouse IgG FITC used 1:200. All conjugates were from Sigma and dilutions employed were as recommended by the manufacturer). The assay was performed using the manufacturer's instructions and cut-offs and a screening dilution of 1:80. A positive reaction was one in which there was distinct fluorescence for >50% of C. burnetii present on the slide well under examination. Initially a positive human sera control supplied with Vircell ELISA was used as a positive control. However, four sheep samples subsequently titrated serially positive to 1:320 and these were employed as positive sheep controls. Positive human sera (with anti-human conjugate) were used as positive control for other animal studies.
RESULTS
Bovine results
A total of 5182 animals (from 273 herds) were tested, 35·4% of which were Friesians and 98·3% female; median age was 4·4 years (quartiles 2·4 and 7·2 years). In total, 2356 (45·5%) animals were from 121 dairy herds, the remainder from beef herds. A total of 330 (6·2%) animals and 132 (48·4%) herds tested positive.
Table 1 summarizes the seroprevalence by herd type (dairy and beef) and by herd size. Seroprevalence was highest (12·5%) in large dairy herds and lowest (2·1%) in small beef herds.
Table 1. Seroprevalence at herd and animal level by herd type (dairy and beef) and size
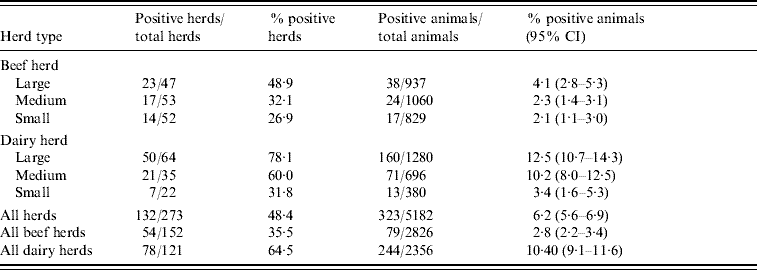
CI, Confidence interval.
Table 2 shows the result of multilevel modelling. Only factors that remained statistically significant (P<0·05) when included in the fully adjusted model are shown in Table 2 (with the exception of sex and Friesian breed).
Table 2. Relationship between animal and herd level characteristics and animal seropositivity
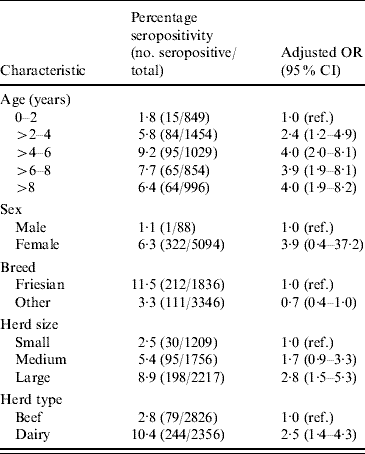
OR, Odds ratio; CI, confidence interval.
Age and breed of animal were related to seropositivity. Animals aged >2 years and >4 years had about twice and 3·5 times, respectively, the odds of seropositivity of animals aged <2 years. There was no further increase in the odds of infection beyond 4 years of age. Friesian animals had the highest seropositivity rate (11·5%). After adjustment for other variables there was some evidence, although of borderline significance, that Friesians were at a higher risk of seropositivity. Animals from dairy herds were twice as likely to be seropositive as animals from other herds and small herds had <40% of the odds of seropositivity of large herds.
Seroprevalence data at both herd and animal level was plotted by geographical region (DVO region) in Figure 1 (animal level) and Figure 2 (herd level). This showed marked variation in seroprevalence between different geographical areas of Northern Ireland. However, when the geographical terms were entered into the model (DVO region and county) these were not related to seropositivity on adjustment for animal and herd characteristics suggesting that the geographic distribution of seropositivity seen in Figures 1 and 2 is explicable in terms of the age and breed of animals and herd size and type.
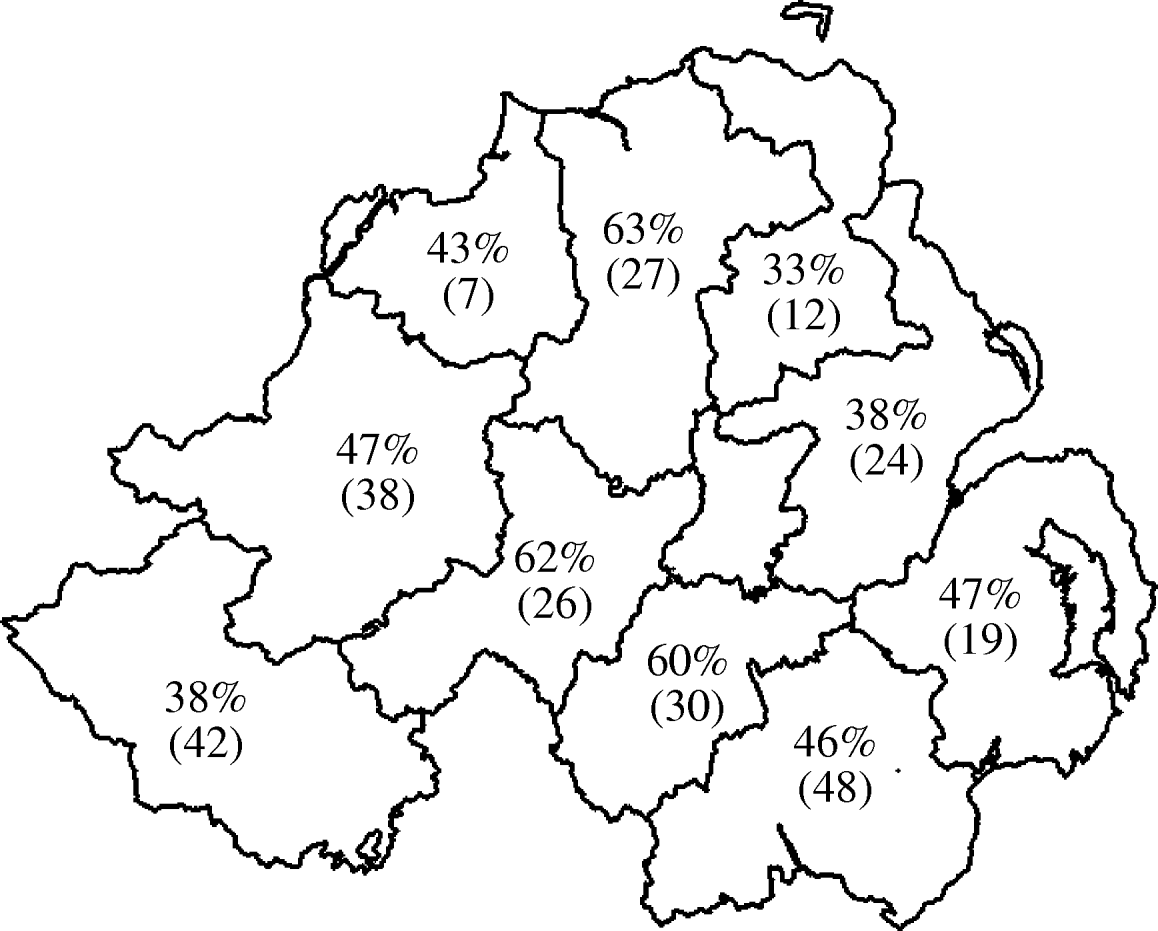
Fig. 2. Geographical distribution of seroprevalence at herd level. Values shown are percent of herds positive with number of herds tested in parentheses.
Bovine validation data for the Bommeli Chekit indirect ELISA
The Bommeli Chekit indirect ELISA used for the survey was validated using the panel of 83 bovine sera. Comparison of the assay with the bioMérieux IFA results are presented in Table 3. Including equivocals by both assays (and regarding these as negative) the calculated sensitivity of the Bommeli Chekit indirect ELISA was 96·3% and the specificity was 96·3%. When the equivocal results with either assay were excluded the sensitivity was 100% and the specificity was also 100%.
Table 3. Bommeli Chekit indirect ELISA kit compared with bioMérieux indirect immunofluorescence (IFA) kit for the detection of Q fever phase 2 IgG antibodies in bovine sera
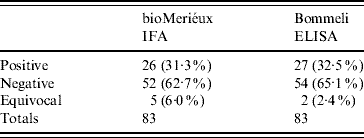
Including equivocals: sensitivity=96·3%, specificity=96·3%.
Excluding equivocals: sensitivity=100%, specificity=100%.
Seroprevalence in ovine, caprine, porcine, rat and mouse samples
Regarding the ovine seroprevalence, 1022 sheep were tested from 58 individual flocks. The ovine seroprevalence was 12·3% at the animal level and 62·1% at flock level. Regarding the caprine study 54 animals from seven flocks were tested. The caprine seroprevalence was 9·3% at the animal level and 42·9% at the flock level. The rodent seroprevalence was 9·7% (3/31) for rats and 3·2 (1/31) for mice.
DISCUSSION
This study utilized detection of IgG to phase 2 in animals. Presence of such antibody indicates past exposure and is not evidence of current infection or shedding of bacteria as animals may seroconvert without detectable shedding and can remain seropositive years after infection has resolved [Reference McQuiston, Childs and Thompson25]. In addition, it is clear that some animals which shed bacteria never seroconvert [Reference McQuiston, Childs and Thompson25].
Recent outbreaks in The Netherlands, Germany and Denmark, apparently associated with small ruminants, have heightened awareness of Q fever as an important zoonoses [Reference Schimmer26]. The data presented in our study are further testament to the ubiquity of infection with this zoonotic agent in ruminant populations. The data suggest somewhat higher bovine seroprevalence rates than England and Wales with a dairy herd seroprevalence of 64·5% in Northern Ireland compared to 21% of herds in England and Wales assessed by antibody testing of bulk milk samples [Reference Paiba15]. However, the methodologies are very different and therefore make comparisons difficult.
Any samples that demonstrated any reactivity on the initial Brucella screening test and SAT, were unavailable to test (2–3% of the samples). This might prompt concerns about bias in sample selection. However Brucella testing process involves an initial SAT test at 1/10 dilution, without EDTA as a sensitive and non-specific screening test. This precautionary approach identifies non-specific reactions in up to 3% of samples. More than 99% of such initial reactive samples prove Brucella negative after confirmatory testing by SAT titration and complement fixation test. We feel that these non-specific SAT tests are of no biological significance. They are seen in all herd types and animal breeds and age groups and from all regions and are unlikely to have any biologically plausible association with Q fever infection that would bias the point-prevalence estimates.
These data clearly indicate that older animals and animals of Friesian breed have increased odds of having been infected. Moreover, these odds are further increased if the animal comes from a large herd, especially if it is a dairy herd. The geographical variables entered into the fully adjusted model (DVO region and county) were not related to seropositivity after adjustment for animal and herd characteristics. Since geographic analysis did not reveal any significant regional differences this suggests that the observed marked geographical distribution of seropositivity is explainable in terms of age and breed of animals and herd size and type.
Age of acquisition data is clearly indicative of horizontal rather than vertical transmission. It is interesting that this appears to plateau beyond year 3 of life. Interestingly, first parturition for the majority of cows in Northern Ireland occurs between ages 2 and 3 years. It is therefore postulated that if a herd is infected with C. burnetii, then the maintenance of infection is mainly due to circulation of the agent within the adult herd. This seems to concur with previous knowledge on the transmission of Q fever occurring around the time of parturition [Reference Parker, Barralet and Bell2, Reference Paiba15]. These data will be useful for modelling potential effectiveness of vaccine strategies being investigated for use in cattle [Reference Guatteo27].
The geographical distribution of cattle seroprevalence bears little relationship to the geographical distribution of human clinical cases in Northern Ireland. Diagnosed human cases mainly occur in upland regions such as North Antrim, the Sperrins and the Mourne mountains where sheep and beef suckler farming predominates. It may be that the relationship between recognized clinical illness in humans and cattle seroprevalence is complex and that clinical cases might be more frequent in areas where susceptibility is highest and seroprevalence lowest. These data suggest that dairy farmers should be the occupational group most exposed to Q fever as they have daily contact with high-prevalence herds including close contact with animals during and after parturition. A seroprevalence study of farmers to investigate the relative risks for differing types of farming practice including both sheep and dairy farmers is underway to test this hypothesis.
The observed ovine and caprine seroprevalence at animal and herd level is similar to that described in the literature for a wide variety of geographical areas [Reference Parker, Barralet and Bell2]. Although the numbers of samples tested and the geographical range were fairly limited in the current study, rat seroprevalence is rather lower than previously reported for England [Reference Webster, Lloyd and Macdonald28]. Regarding the lack of pig seroprevalence, this suggests that pigs are not important in the ecology of Q fever. There is very little previous information in the literature regarding Q fever infection of pigs.
This is the first bovine serological survey to have been carried out in Northern Ireland and the most comprehensive to be carried out in Europe. It has demonstrated a relatively high prevalence of current or past infection in the cattle population which has clear zoonotic implications. Without studies like this, understanding of the transmission and maintenance of C. burnetii in ruminants cannot be advanced and the public health implications of particular activities or changes in animal husbandry cannot be understood. Studies are continuing to further elucidate the ecology of Q fever infection in Northern Ireland.
ACKNOWLEDGEMENTS
This study was funded by a grant from the Research & Development Office, a directorate of the Northern Ireland Health and Social Services Central Services Agency (RRG project 9.8). The authors are grateful to Charles Torbitt for the production of the maps used in this paper.
DECLARATION OF INTEREST
None.







