Introduction
Plant hybridization is a common process in nature, and plays an important role in plant breeding. Hybridization can generate a series of novel phenotypes, including a broad array of new, and sometimes transgressive, phenotypes (Rieseberg et al., Reference Rieseberg, Archer and Wayne1999). It can also result in speciation, adaptive evolution and ecological innovations (Rieseberg, Reference Rieseberg1997; Rieseberg et al., Reference Rieseberg, Raymond, Rosenthal, Zhao, Livingstone, Nakazato, Durphy, Schwarzbach, Donovan and Lexer2003; Arnold, Reference Arnold2004; Hegarty & Hiscock, Reference Hegarty and Hiscock2005). Inter-specific crosses in plants often generate hybrids that exhibit heterosis compared with their parents, which provides a vast reservoir of new alleles for gene evolution (Zhuang & Adams, Reference Zhuang and Adams2007). Allelic variation resulting from hybridization can contribute to phenotypic variation. For example, the complementation and interaction of different alleles in hybrids are hypothesized to be a component of the genetic basis for heterotic phenotypes (Birchler et al., Reference Birchler, Auger and Riddle2003; Guo et al., Reference Guo, Rupe, Zinselmeier, Habben, Bowen and Smith2004; Song et al., Reference Song, Segal and Messing2004; Birchler et al., Reference Birchler, Yao and Chudalayandi2006; Springer & Stupar, Reference Springer and Stupar2007).
Allelic expression differences of genes have also been reported in inter-specific hybrids of Drosophila (Wittkopp et al., Reference Wittkopp, Haerum and Clark2004), as well as intra-specific F1 hybrids of mice (Cowles et al., Reference Cowles, Hirschhorn, Altshuler and Lander2002) and Saccharomyces cerevisiae (Ronald et al., Reference Ronald, Brem, Whittle and Kruglyak2005). Allelic variation is often attributed to qualitative changes that affect the nature of the gene products, and to quantitative changes that alter their level of expression (Stupar & Springer, Reference Stupar and Springer2006). Quantitative changes in allele expression may be the result of variation by regulatory factors (Wittkopp et al., Reference Wittkopp, Haerum and Clark2004). Rockman & Kruglyak (Reference Rockman and Kruglyak2006) define the two types of regulatory sequence variation as local and distant regulatory variation. Local variation, which maps close to the physical location of the affected gene, influence transcription in an allele-specific manner; distant variation, located elsewhere in the genome, acts in trans through the downstream effects of coding or cis-regulatory polymorphisms in different types of genes (Rockman & Kruglyak, Reference Rockman and Kruglyak2006).
Some studies have indicated that much of the allelic expression variation might be attributed to local variation. For example, as many as 25% of all gene expression traits in a yeast cross are affected by local regulatory variation (Ronald et al., Reference Ronald, Brem, Whittle and Kruglyak2005). Stupar & Springer (Reference Stupar and Springer2006) classified 18 of 35 maize allelic genes as local variation. Zhuang & Adams (Reference Zhuang and Adams2007) classified 6 of 19 Populus F1 hybrid allelic genes as local variation. However, no cis-elements or transcription factors that could correlate with allelic expression variation were mentioned in detail.
In previous studies, we detected allelic expression variation in the leucine-rich receptor-like kinase (LRK) gene cluster, in which alleles of some genes were unequally expressed in hybrids of Nipponbare (Oryza sativa L. subsp. Japonica var. Nipponbare)/93-11 (O. sativa L. subsp. Indica var. 93-11) cross (He et al., Reference He, Luo, Tian, Li, Zhu, Su, Qian, Fu, Wang, Sun and Yang2006). Here, we studied allelic variation in gene expression levels of the yield-related quantitative gene, LRK6, using the parental species and the F1 hybrid. To determine if biased allelic expression was the result of hybridization or reflected differing expression levels in the parents, we compared the ratio of specific transcripts in the hybrid and its parents. To investigate the regulation of allele expression, we isolated and analysed the promoter of LRK6 by successive 5′ deletions. We cloned a rice ethylene-responsive factor (ERF) gene, O. sativa ERF3 (OsERF3), via a yeast one-hybrid system. OsERF3 binds to a newly identified motif in the LRK6 promoter, which might be responsible for the differential expression of LRK6 in 93-11 and Nipponbare.
Materials and methods
Plant material and allele expression analysis of LRK6
Plant varieties used were rice var. 93-11, var. Nipponbare and the Nipponbare/93-11 hybrid. DNA was isolated from fresh leaves for testing the amplification efficiency of primers. Total RNA was isolated from fresh leaves at the three-leaf stage using TRIzol (Gibco BRL, USA), and then treated with RNase-free DNase I (Sigma, USA) to reduce DNA contamination. Poly(A)+ mRNA was purified by a Poly(A) Tract kit (Promega, USA) and used for reverse transcription using SuperScript™ II RNase H− reverse transcriptase (Invitrogen, USA). cDNA was then amplified by PCR using primer1-F (5′-GGACATTTTTAATAAAATTTTGTGG-3′), primer1-R (5′-GGAGATGAAATCAGAAGGGAAT-3′), primer2-F (5′-TCATCAGGTCCATTTCATTTG-3′) and primer2-R (5′-ACAGCTTTTTTTTTTGTACAGCTT-3′). Primer1 was specific to Nipponbare and Primer2 was specific to 93-11. The rice actin1 gene (NCBI accession No. X16280.1) was amplified using primers actin-F (5′-CTGTCTTCCCCAGCATTGTC-3′) and actin-R (5′-GGTCTTGGCAGTCTCCATTTC-3′) to serve as a positive control for quantification of the relative amounts of cDNA. The semi-quantitative reverse transcription (RT)-PCR analysis followed the protocols of He et al. (Reference He, Luo, Tian, Li, Zhu, Su, Qian, Fu, Wang, Sun and Yang2006) .
Isolation of the LRK6 promoter
The 5′-flanking region of LRK6 was isolated from rice var. 93-11 genomic DNA. The primers were designed from the LRK cDNA sequence (NCBI accession No. AY730046) and total genomic DNA was used as a template. The primers were LRK6P-F (5′-GAGGAAAATATCAAAACGACT-3′) and LRK6P-R (5′-CATGGCCTCCAAGCAAAT-3′). PCR cycling conditions were 5 min at 94°C; 30 cycles of 30 s at 94°C, 30 s at 64°C and 1 min at 72°C; and 10 min at 72°C for the final extension step. The LRK6 promoter was directly amplified as a 1466 bp fragment from rice genomic DNA and cloned into pMD19-T (TaKaRa, Japan) for sequencing.
Preparation of LRK6 promoter deletions
A series of 5′ promoter deletions were generated by PCR using the reverse primer LRK6P+1-R (5′-GGCCTCCAAGCAAATGGT-3′) and the following forward primers: LRK6P-F; LRK6P-1366-F (5′-AAACAAAATCAAAACATCCTAC-3′); LRK6P-1263-F (5′-CAGCGGAGATGAGCCAAGG-3′); LRK6P-1190-F (5′-TCAGACTTTCAGTGGCATAG-3′); LRK6P-1089-F (5′-ATTGTCGAACCATTTCCG-3′); LRK6P-948-F (5′-TCCAGACGCAGGATGAAA-3′); LRK6P-868-F (5′-GCTATAGCTTTGGCGTCT-3′); LRK6P-766-F (5′-TTAGCGACTAACAAGTAATG-3′); LRK6P-620-F (5′-CCGATTTGTTCTGGGATA-3′); and LRK6P-518-F (5′-TGGGAATGACCAACACTG-3′). The annealing temperatures for PCR cycling using the above primers were 64, 58, 62, 60, 60, 63, 61, 60, 59 and 59°C, respectively. The deletion fragments, containing terminal HindIII and BglII sites, were cloned into pMD19-T. All deletion fragments were confirmed by sequencing.
Construction of plant transformation vectors
The binary plasmid vector pCAMBIA1304 (Centre for the Application of Molecular Biology to International Agriculture (CAMBIA), Canberra, ACT, Australia), which carries a kanamycin resistance gene for bacterial selection and a hygromycin phosphotransferase gene (hyp) for plant transformation selection, was used. Oryza sativa L. programmed cell death 5 (OsPDCD5) plays an essential role in cell death in rice plants and causes a number of morphological changes in transgenic plants (Attia et al., Reference Attia, Li, Wei, He, Su and Yang2005). We therefore chose this gene as a reporter for the promoter deletion study. A control binary plasmid was constructed by inserting the OsPDCD5 cDNA in the sense orientation, driven by the cauliflower mosaic virus (CaMV) 35 s promoter and the Nos-3′ terminator, between the Bg1II and SpeI sites in pCAMBIA 1304. Deletions in pMD19-T were excised with HindIII and BglII and cloned into pCAMBIA1304. The deletion fragments were substituted for the CaMV 35 s promoter of the expression vector. This generated a series of constructs containing an OsPDCD5 expression cassette regulated by an LRK6 promoter deletion. Deletions were numbered in the 5′ direction from the first nucleotide in the LRK6 start codon (defined as +1). The full-length LRK6P::OsPDCD5 reporter construct was designated as pCAMBIA1304-OsPDCD5-1465 (abbreviated to p-1465). Deletions were designated as p-1366, p-1263, p-1190, p-1089, p-948, p-868, p-766, p-620 and p-518.
Plant transformation
Mature seeds of rice var. 93-11 were husked and surface sterilized by immersion in 70% ethanol for 2 min, followed by washing in sterile distilled water. Seeds were then soaked in 0·1% HgCl2 for 20 min with regular shaking, rinsed with several changes of sterile distilled water, dried on sterilized filter papers and inoculated on medium for embryogenesis callus (Attia et al., Reference Attia, Li, Wei, He, Su and Yang2005). We selected about 30 pieces per dish and bombarded them twice (Attia et al., Reference Attia, Li, Wei, He, Su and Yang2005).
Yeast one-hybrid screening
The yeast one-hybrid assay was performed using a MatchMaker One-Hybrid Library Construction & Screening kit supplied by Clontech (TaKaRa, Japan). Rice cDNA library construction, yeast culture techniques and yeast transformation were performed as described in the MatchMaker One-Hybrid Library Construction & Screening Manual (Clontech No. PT3529-1) and Yeast Protocols Handbook (Clontech No. PT3024-1). The full sequence of differential sequence of LRK6 promoter 2 (DSLP2) (5′-CTCTAAAGTTAAGT-3′) from the rice LRK6 promoter was synthesized into three tandemly repeated copies and then inserted into the EcoRI–MluI sites of the multiple cloning site (MCS) upstream of the HIS3 minimal promoter in the pHIS2.1 expression vector (TaKaRa, Japan). Total RNA was isolated from leaves and roots of 93-11 using RNA prep pure Plant Kit (TIANGEN, Beijing, China). The protocol of cDNA library construction followed the manufacturer's manual. Rice cDNAs were fused into a region downstream of the transcriptional activation domain of the yeast expression vector pGADT7-Rec2, by homologous recombination. The prey and bait plasmids were introduced into yeast strain Y187. Transformants were first selected using selective medium (without tryptophan, leucine and histidine) containing 5 mM 3-AT. After confirmation of positive interaction, prey plasmids were isolated using a yeast plasmid kit (Biomiga, San Diego, USA), transformed into Escherichia coli DH5α and sequenced.
Identification of the cis-element
According to analysis of DSLP2 by the PLACE web signal scan program (http://www.dna.affrc.go.jp/PLACE/signalup.html), DSLP2 was divided into three parts: DSLP2-1 (5′-TACT-3′), DSLP2-2 (5′-CTAAAGT-3′) and DSLP2-3 (5′-TAAGT-3′). Using DSLP2-3 as a template, fragments were prepared by substituting T with G and G with T, and named DSLP2-4 (5′-GAAGT-3′), DSLP2-5 (5′-TAATT-3′) and DSLP2-6 (5′-TAAGG-3′). All fragments were synthesized into four tandem copies and inserted into the pHIS2.1 vector for the one-hybrid assay. The interaction between the transcription factor and DNA sequence was tested by growth on media lacking Trp, Leu and His. His synthase inhibitor 3-AT (5 mM) was added to the media to suppress background activation and assess the strength of the interaction.
GCC-binding assay via yeast one-hybrid screen
ERFs contain a highly conserved DNA-binding domain known as the ERF domain, and were first identified as transcription factors positioned downstream of the ethylene signalling pathway. ERFs modulate the expression of many downstream genes through the GCC-box present in their promoters (Ohme-Takagi & Shinshi, Reference Ohme-Takagi and Shinshi1995; Sessa et al., Reference Sessa, Meller and Fluhr1995; Shinshi et al., Reference Shinshi, Usami and Ohme-Takagi1995). The DNA binding ability of OsERF3's ERF/AP2 domain was analysed using the one-hybrid assay. Both the wild-type GCC fragment, containing three copies of the GCC box sequence (GCC: 5′-TAAGAGCCGCC-3′) and the mutant mGCC fragment, containing three copies of the mutated GCC box sequence (mGCC: 5′-TAAGATCCTCC-3′), were synthesized and prepared following the protocols of Ohme-Takagi & Shinshi (Reference Ohme-Takagi and Shinshi1995) and Mazarel et al. (Reference Mazarel, Puthoff, Hart, Rodermel and Baum2002) .
Ethephon treatments in 93-11 and Nipponbare
O. sativa L. subsp. Indica var. 93-11 and O. sativa L. subsp. Japonica var. Nipponbare seeds were used in all experiments. Seeds were germinated at 37°C in the dark for 2 days and then transferred to a plant growth chamber to grow to the three-leaf stage under controlled conditions (12 h light/12 h dark 26°C cycle). For hormone treatments, seedlings at the three-leaf stage were incubated in 100 μM ethephon solution (ethylene) for 1, 3, 6, 12 and 24 h at 26°C, respectively. RNA samples were isolated from leaves for marker gene analysis. Untreated seedlings were used as a control.
Quantitative RT-PCR of OsERF3
Total RNAs were extracted from all samples using RNAiso Plus (TaKaRa), according to the manufacturer's instructions. The RNA was reverse-transcribed using PrimeScript RT reagent kit (TaKaRa). cDNAs were specifically amplified with the following sets of primers: ERF3-F (5′-GCCGACTCTGGACTTGGATTTGTTC-3′) and ERF3-R (5′-TGCCGCCTTGTTCGCCGTAA-3′); LRK6-F (5′-CGGCAATCTTAGCAATGTGA-3′) and LRK6-R (5′-GATAACCGAAGTGCGACCA-3′). Quantitative PCR was performed on a Bio-Rad real time PCR system using SYBR Premix Ex Taq™ (TaKaRa). The PCR thermal cycle conditions were as follows: denaturation at 95°C for 30 s; followed by 40 cycles of 95°C, for 5 s and 60°C for 30 s. The rice OsActin1 gene, which was amplified with primers Actin-F (5′-TCTGGCATCATACCTTCTACA-3′) and Actin-R (5′-GGATGGCTGGAAGAGGAC-3′), was used as an internal reference gene for calculating relative transcript levels.
Results
Allele-specific expression of LRK6 in hybrids
In previous studies, the expression of LRK6 was demonstrated to be constitutive and the 5′ and 3′ untranslated sequences of LRK6 were compared in detail between Nipponbare and 93-11 (He et al., Reference He, Luo, Tian, Li, Zhu, Su, Qian, Fu, Wang, Sun and Yang2006). We found two allelic sequence polymorphisms located in the untranslated region. A 23 bp indel appeared to be a deletion in 93-11 relative to Nipponbare in the 5′ untranslated region of LRK6, and a 17 bp indel appeared to be a deletion in Nipponbare relative to 93-11 in the 3′ untranslated region. Using these polymorphisms, allele-specific primers were designed for RT-PCR analysis of Nipponbare, 93-11 and the Nipponbare/93-11 hybrid (Fig. 1(a)). The equivalent DNAs of Nipponbare and 93-11 were amplified by primer1 and primer2. Primer1 was specific for amplification in Nipponbare, and primer2 was specific for amplification in 93-11. There was no obvious difference in amplification efficiency of two pairs of primers in Nipponbare and 93-11 (Fig. 1(b)). For cDNA amplification, both parental alleles of LRK6 were expressed, but they did not contribute equally to the total amount of transcript in the hybrid. In the parental lines, the Nipponbare allele was expressed about 2·18× higher level than the 93-11 allele. In the hybrid, the expression levels of the Nipponbare and 93-11 alleles were similar to that of each parent, respectively. The expression level of Nipponbare allele was about 2·5× higher than that of the 93-11 allele, which was consistent with the result of previous work (He et al., Reference He, Luo, Tian, Li, Zhu, Su, Qian, Fu, Wang, Sun and Yang2006) (Fig. 1(c)).
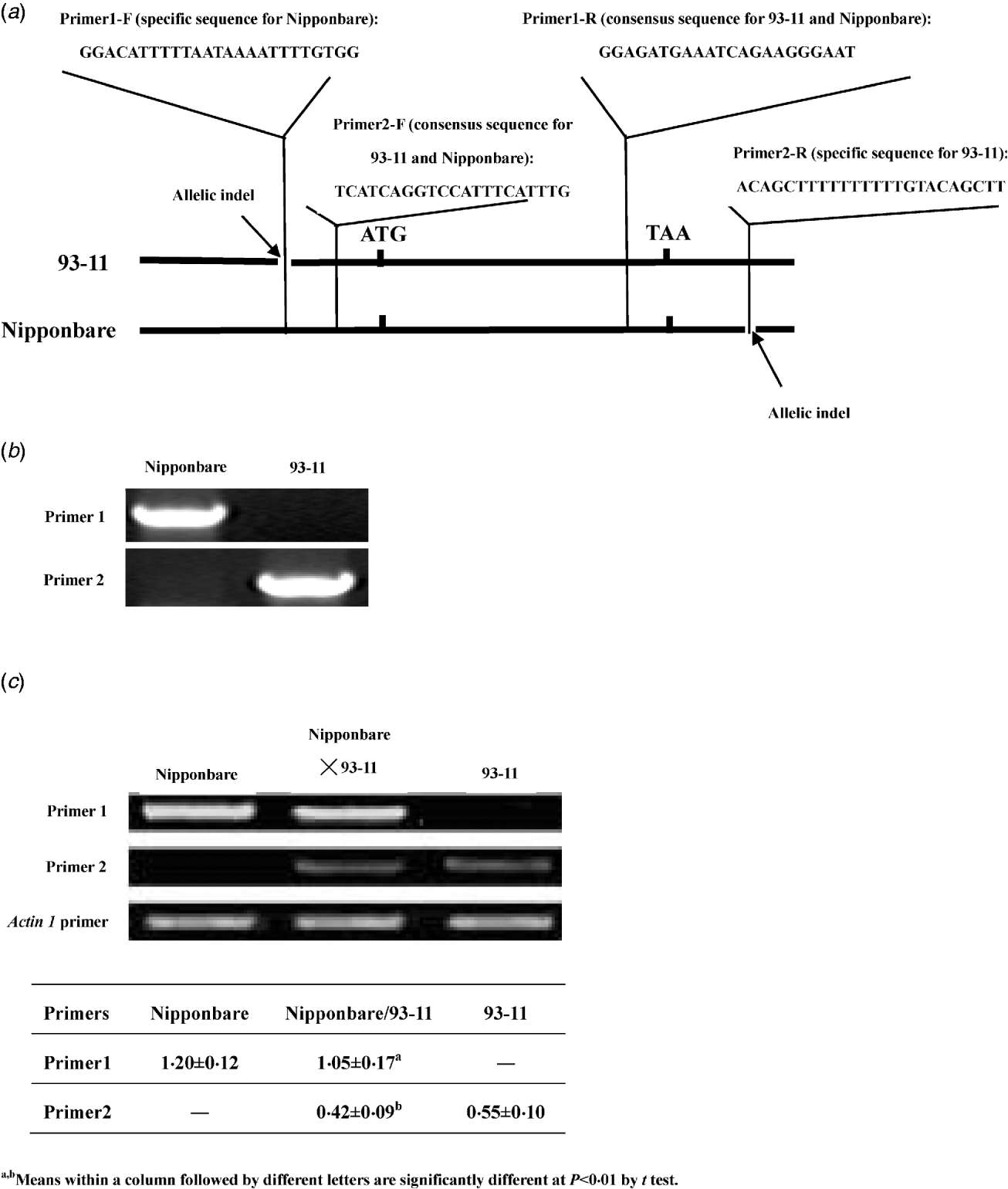
Fig. 1. Allele-specific expression assay of LRK6. (a) Schematic diagram of primer design for allelic-specific expression analysis of LRK6. Two small allelic indels in the 5′ and 3′ non-coding regions of LRK6 were used to design the allelic-specific primers. (b) Amplification efficiency test of primer1 and primer2 in Nipponbare and 93-11. (c) Allele-specific expression of LRK6 in Nipponbare, 93-11, and the Nipponbare/93-11 hybrid. Rice actin1 was used as a control.
Isolation and analysis of the LRK6 promoter
We analysed the expression level of each individual LRK gene in Nipponbare, 93-11, and the Nipponbare/93-11 hybrid by RT-PCR. As noted above, LRK6 was expressed at a much higher level in Nipponbare than in 93-11, which is probably the result of sequence variation in the regulatory regions of the promoters. We isolated about 1·5 kb of the 5′-flanking region of LRK6 from var. 93-11 by PCR, and divided the 5′ upstream region of LRK6 into ten segments for deletion experiments (Delaney et al., Reference Delaney, Orford, Martin-Harris and Timmis2007). Successive 5′ deletions of the LRK6 promoter were linked to an OsPDCD5 reporter gene in pCAMBIA1304 constructs, which were bombarded into a variety of rice calli. We identified a specific region in the LRK6 promoter that was essential for OsPDCD5 reporter gene expression (Fig. 2).
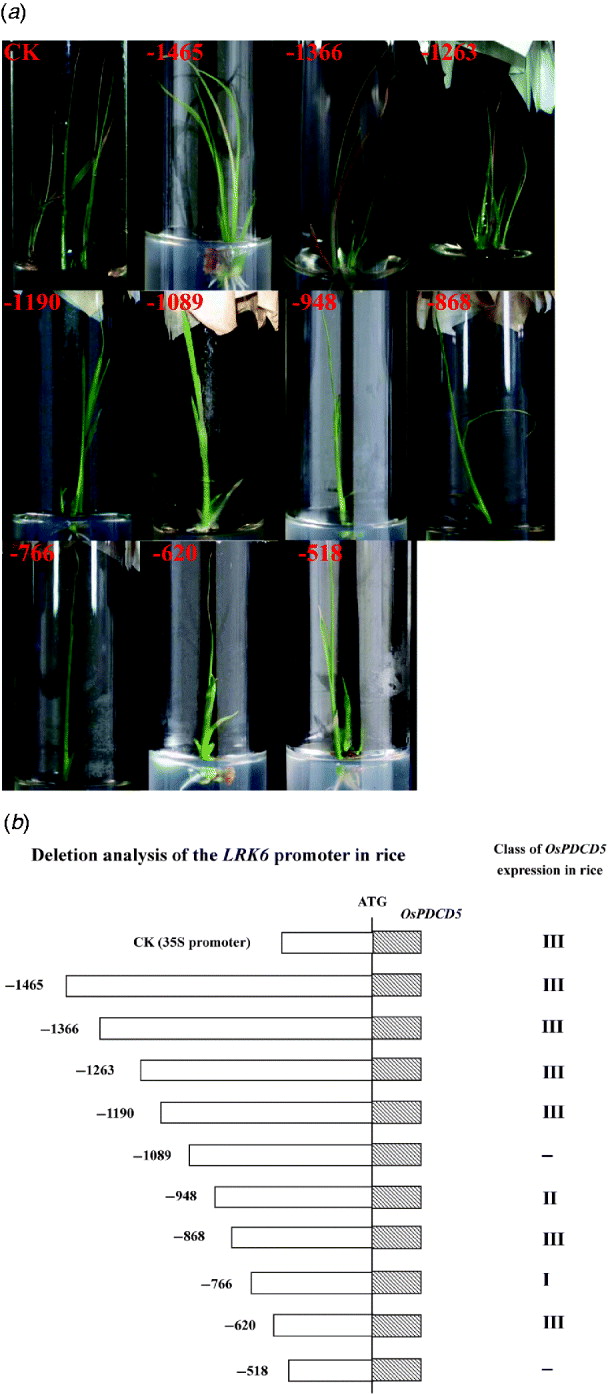
Fig. 2. Activity of the LRK6 promoter in transgenic rice. (a) Rice calli were bombarded with reporter plasmids containing OsPDCD5 regulated by successive deletions of the LRK6 promoter. The bombarded tissues were allowed to differentiate for at least 1 month. Transgenic seedlings had typical programmed cell death phenotypes. CK, 35S::OsPDCD5; 1, p-1465; 2, p-1366; 3, p-1263; 4, p-1190; 5, p-1089; 6, p-948; 7, p-868; 8, p-766; 9, p-620; 10, p-518. (b) Based on the effect of OsPDCD5 expression on the morphology of transgenic rice, CK (35S::OsPDCD5) induced morphological features of cell death, including leaf yellowing, early leaf senescence, growth inhibition and early death, as did p-1465, p-1366, p-1263, p-1190, p-868 and p-620. Transgenic seedlings of p-1089 and p-518 did not show any cell death phenotypes; p-948 displayed leaf yellowing and early leaf senescence; p-766 displayed only early leaf senescence.
Our previous studies in plants indicated that OsPDCD5 activity induced morphological features of cell death in transgenic plants, including precocious induction of leaf yellowing, early leaf senescence, growth inhibition and early death (Attia et al., Reference Attia, Li, Wei, He, Su and Yang2005). For standardization, we divided the morphological changes into three classes. Class I was only leaf yellowing. Class II had leaf yellowing, early leaf senescence and growth inhibition. Class III displayed all morphological features of cell death. If there were more than three transgenic plants showing the same phenotype, then that phenotype defined the class to which they belonged. Bombardment of rice calli with an OsPDCD5 reporter construct regulated by the CaMV 35S promoter and LRK6 promoter produced morphological changes in transgenic rice. The control 35S::OsPDCD5 construct showed a clear phenotype of cell death in transgenic rice, as did p-1465, p-1366, p-1263, p-1190, p-868 and p-620. Thus, these deletions belonged to class III. Interestingly, the p-1089 and p-518 constructs did not cause cell death, while there was a slight morphological change in plants carrying the p-948 and the p-766, which belonged to class II and class I, respectively (Fig. 2). Based on the deletions of the LRK6 promoter, two regions were identified in the promoter sequence that affected the expression of LRK6.
The 1465 bp LRK6 promoter, and the deletions down to −1190, displayed clear signs of cell death, demonstrating that the promoter region from −1465 to −1190 is sufficient for gene expression. The deletion from −1089 to −948 did not cause cell death, indicating that this promoter region might have elements essential for restricting gene expression in rice. The deletion from −766 to −620 produced a slight form of cell death, suggesting that this region has binding sites for transcription factors. Deletion to −518 abolished OsPDCD5 expression in all seedlings tested, showing that the region from −518 has lost all elements necessary for promoter function (Fig. 2(b)).
We generated constructs to define the promoter regions important for gene expression and minimal promoter activity more accurately. The deletion from −1089 to −948 and from −766 to −620 directed differential OsPDCD5 expression levels in transgenic rice (Fig. 2(a)). This localized the elements necessary for expression to two ~100 bp regions of the LRK6 promoter. These regions appeared to contain elements that changed the promoter activity. Interestingly, there were two gaps in the sequence of the LRK6 promoter between 93-11 and Nipponbare, when compared by DNAssist software (Fig. 3). These differences in promoter sequence were named DSLP1 and DSLP2. DSLP1 (−1089 to −948 of the LRK6 promoter) is 5′-TA-3′ in 93-11 and 5′-CAACAA-3′ in Nipponbare (Fig. 3(a)); while DSLP2 (−766 to −620) is 5′-CTCTAAAGTTAAGT-3′ in 93-11, and completely absent from Nipponbare (Fig. 3(b)). PLACE analysis showed that DSLP2 had potential transcription factor binding sites; therefore, we used yeast one-hybrid assays to identify proteins that might cause the differential expression of LRK6 in 93-11 and Nipponbare.

Fig. 3. Sequences of the LRK6 promoter were compared in 93-11 and Nipponbare. (a) The sequence from −1089 to −948 of the LRK6 promoter differed in the two varieties; 5′-CAACAA-3′ in Nipponbare, and 5′-TA-3′ in 93-11. (b) The sequence from −766 to −620 of the LRK6 promoter showed discrepancies in Nipponbare and 93-11.
Characterization of an AP2/ERF protein that interacts with the DSLP2 segment
A yeast one-hybrid assay was used to isolate rice 93-11 proteins that interact with DSLP2. We generated a yeast strain containing three direct tandem repeats of DSLP2 regulating the HIS3 gene and transformed it with a cDNA library consisting of pGADT7-Rec2 fused to the GAL4 activation domain. Approximately 3·75×106 clones were screened, and 48 HIS3-positive clones were isolated after three rounds of selection. Plasmid rescue and cDNA sequencing identified three identical cDNA clones encoding an AP2/ERF protein, designated rice ERF3 (OsERF3). Yeast clones containing OsERF3–GAL4 consistently demonstrated strong growth on 5 mM 3-amino-1,2,4-triazole (3-AT) in repeated experiments. The OsERF3 cDNA sequence encoded a complete open reading frame consisting of 708 nucleotides with no introns (Fig. 4(a)).

Fig. 4. OsERF3 gene sequence, protein sequence and protein domains. (a) The OsERF3 gene and its protein sequence. OsERF3 encodes a complete open reading frame of 236 amino acids with a predicted molecular mass of 24·3 kDa. (b) Alignment of the N-terminal and C-terminal sequences of the rice ERF protein (Ohta et al., Reference Ohta, Matsui, Hiratsu, Shinshi and Ohme-Takagi2001), Arabidopsis (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000) and tobacco (Ohta et al., Reference Ohta, Ohme-Takagi and Shinshi2000) indicates that there are conserved ERF and EAR motifs in OsERF3.
The OsERF3 gene was predicted to encode a protein of 235 amino acids, with a calculated molecular weight of 24·3 kDa. OsERF3 is an AP2/ERF protein with an N-terminal domain containing one highly conserved ERF motif, and a C-terminus containing an ~30 amino acid domain comprising a conserved ERF-associated amphiphilic repression (EAR) motif (Fig. 4(b)). The large ERF family of transcription factors is part of the AP2/ERF superfamily (Riechmann et al., Reference Riechmann, Heard, Martin, Reuber, Jiang, Keddie, Adam, Pineda, Ratcliffe, Samaha, Creelman, Pilgrim, Broun, Zhang, Ghandehari, Sherman and Yu2000), and is further divided into two major subfamilies: ERF and CBF/DREB (C-repeat binding factor/Dehydration responsive element binding protein) (Sakuma et al., Reference Sakuma, Liu, Dubouzet, Abe, Shinozaki and Yamaguchi-Shinozaki2002). Several genes involved in plant growth and development encode ERF proteins. The EAR motif confers the capacity for repression of a heterologous DNA binding domain, and the motif is essential for such repression (Ohta et al., Reference Ohta, Matsui, Hiratsu, Shinshi and Ohme-Takagi2001).
The EAR motif is also found in proteins from Arabidopsis and tobacco (Fig. 4(b)). The OsERF3 sequence showed 24–44% identity to other identified ERFs, including Arabidopsis AtERF3, 4 and 7–12, and tobacco NtERF3. The ERF domains of the OsERF3 protein show a high level of identity to the corresponding ERF domains in other plants. Alignment and phylogenetic tree analysis revealed that OsERF3 is a Class II plant ERF protein (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000; Tournier et al., Reference Tournier, Sanchez-Ballesta, Jones, Pesquet, Regad, Latche, Pech and Bouzayen2003). OsERF3 has 31–40% overall identity, rising to 81–91% for the ERF domain, with Arabidopsis AtERF3 and AtERF4, tobacco NtERF3 and Nicotiana sylvestris NsERF3.
Identification of a cis-element in the LRK6 promoter by division and mutation analysis
To determine the specific region that interacts with OsERF3, DSLP2 (5′-CTCTAAAGTTAAGT-3′) was divided into three parts: DSLP2-1 (5′-TACT-3′), DSLP2-2 (5′-CTAAAGT-3′) and DSLP2-3 (5′-TAAGT-3′), by analysis of DSLP2 using the PLACE web signal scan program (http://www.dna.affrc.go.jp/PLACE/signalup.html). The three fragments were synthesized into four tandem copies and inserted into pHIS2.1 vector for a one-hybrid assay (Fig. 5(a)). OsERF3 bound DSLP2-2 and DSLP2-3 sequences, but had no interaction with DSLP2-1. DSLP2 acted as a positive control interaction, with similar strength. DSLP2-2 and DSLP2-3 segments contained a similar motif (5′-TAA(A)GT-3′), which potentially interacts with OsERF3 (Fig. 5(b)).
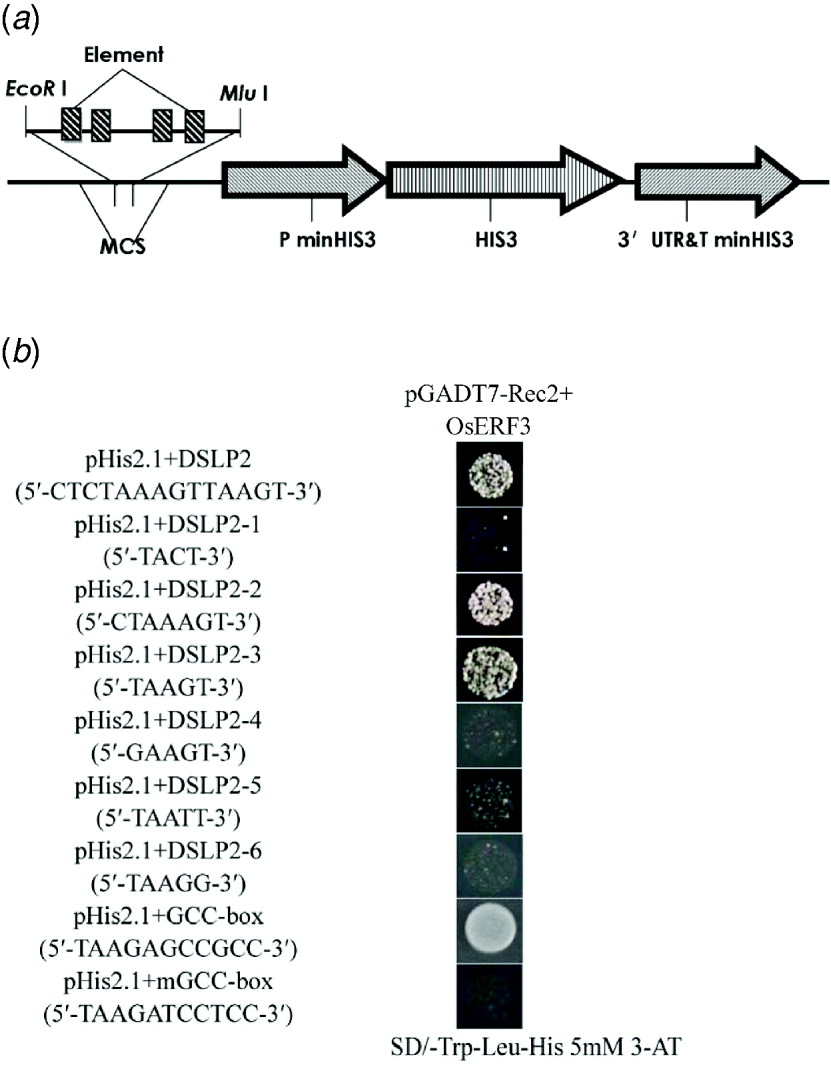
Fig. 5. Identification of a cis-element by division and mutation analysis, and GCC-box binding assay of OsERF3 protein. (a) Structure of the pHIS 2·1 bait construct. The DSLP2 fragment (divisions and mutations of DSLP2) was synthesized into four tandem copies and cloned into the EcoRI and MluI sites of the MCS to increase HIS3 expression. (b) Division analysis in a yeast one-hybrid assay. Bait-reporter constructs containing four copies of DSLP2 fragments, which regulated the expression of HIS3, were transformed into yeast strain Y187 to generate three clones. Positive clones from DSLP2-2 and DSLP2-3 grew on SD/-Trp/-Leu/-His/5 mM 3-AT, whereas no DSLP2-1 clones grew. Wild-type DSLP2 was used as a control. Mutation analysis in an one-hybrid assay. There was a significant decrease in the strength of the interactions for DSLP2-4, DSLP2-5 and DSLP2-6. For GCC-box binding test, transformants of OsERF3 plus the GCC-box had strong binding abilities, but transformants of OsERF3 plus the mutated GCC-box (mGCC-box) had no binding ability.
To further explore whether the TAA(A)GT motif was an effective element for interaction, 5′-TAAGT-3′ was mutagenized by substituting T with G and G with T, to generate DSLP2-4 (5′-GAAGT-3′), DSLP2-5 (5′-TAATT-3′) and DSLP2-6 (5′-TAAGG-3′), and the interaction experiment was repeated. The DSLP2-3 control fragment showed strong interaction, but the strength of the interaction was significantly decreased for DSLP2-4, DSLP2-5 and DSLP2-6 (Fig. 5(b)). The bases T, G and T in this motif therefore play crucial roles in the binding activity of the transcription factor. Together, these results indicate that the TAA(A)GT motif is a crucial cis-element in the LRK6 promoter, which might be responsible for the observed differential expression of LRK6 in 93-11 and Nipponbare.
The OsERF3 protein interacts with the GCC-box in yeast
The ERF/AP2 domain contains key amino acids that bind to GCC-boxes. We therefore overexpressed recombinant pGADT7-Rec2-OsERF3 containing the ERF/AP2 domain in E. coli to examine its GCC binding ability. Purified pGADT7-Rec2-OsERF3 plasmid was mixed with either the pHIS 2·1-GCC-box or the pHIS 2·1-mGCC-box in the binding reaction in a one-hybrid assay (Fig. 5(b)). The GCC-box strongly bound to OsERF3 protein, but the mGCC-box did not. Therefore, the ERF/AP2 domain of OsERF3 can bind to the GCC-box element, but not to the mGCC-box. There was no GCC-box found in the LRK6 promoter of 93-11 and Nipponbare. We concluded that the expression of LRK6 in 93-11 was directly influenced by OsERF3, but in Nipponbare it was not.
Expression profiles of OsERF3 and LRK6 in 93-11 and Nipponbare
To determine the relationship between OsERF3 and LRK6, expression analysis of OsERF3 and LRK6 was performed in 93-11 and Nipponbare by ethylene treatment. Generally, ethephon or 1-aminocyclorpopane-1-carboxylic acid replaced ethylene for the hormone treatments. There were obvious differences of expression of OsERF3 and LRK6 between 93-11 and Nipponbare. The expression level of OsERF3 in both 93-11 and Nipponbare increased dramatically after 1 h of treatment (Fig. 6). The expression of LRK6 in 93-11 and Nipponbare also increased, but the level in Nipponbare was relatively low compared with 93-11. After 3 h of treatment, the expression of OsERF3 in both 93-11 and Nipponbare decreased significantly. The expression of LRK6 decreased more in 93-11 than in Nipponbare. When treatment was continued for more than 3 h, the expression level of OsERF3 and LRK6 tended to become stable, probably because the stress response had disappeared. We found that the expression patterns of OsERF3 and LRK6 in 93-11 were different from those in Nipponbare at 6, 12, and 24 h of treatment. In Nipponbare, the expression of OsERF3 increased after 6 h of treatment, and decreased after 12 and 24 h, as did the expression of LRK6. While the expression of OsERF3 in 93-11 decreased after 6 h of treatment, increased after 12 h and decreased in 24 h, the opposite was true for the expression of LRK6. When the expression of OsERF3 was up-regulated, LRK6 expression decreased in 93-11. These results suggest that OsERF3 acts as a repressor to adjust the expression of LRK6 in 93-11, but has no repressive effect on the expression of LRK6 in Nipponbare. The results of ethephon treatment indicated that the expression of LRK6 could be connected to the plant ethylene response pathway, and that OsERF3 might directly control the expression of LRK6 in 93-11. This might also explain why the alleles of LRK6 were unequally expressed in hybrids of the Nipponbare/93-11 cross.
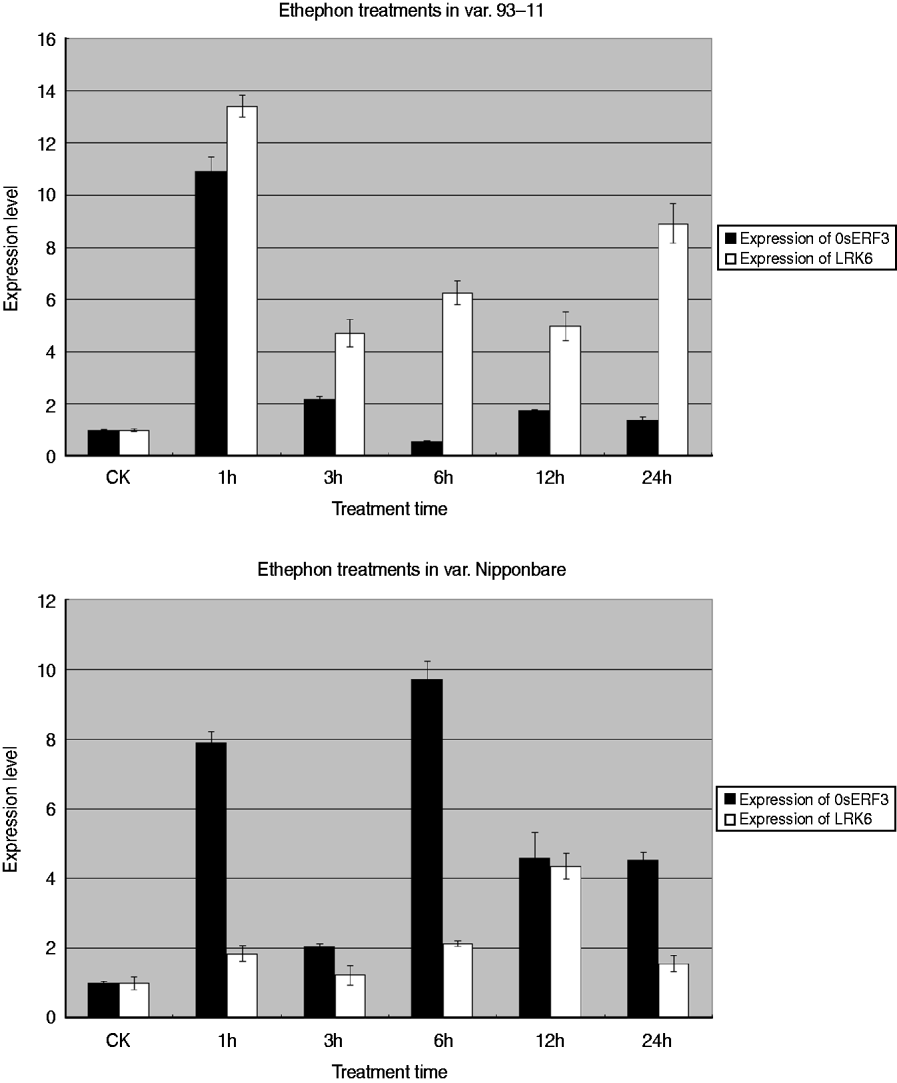
Fig. 6. Real-time-PCR analysis of OsERF3 and LRK6 expression in 93-11 and Nipponbare. Expression patterns and levels of OsERF3 and LRK6 were different in 93-11 and Nipponbare. The durations of the ethephon treatments are indicated. Non-treated seedlings were used as controls, error bars, ±sd.
Discussion
OsERF3, acting as a repressor, may account for the allele-specific expression of LRK6
Many studies demonstrated some important traits affected by a cis-element and its interaction with a repressor in plants. In rice, five novel cis-elements were identified from green tissue-specific promoter PD540 . Two of them, interacted with transcription factor, down-regulated the tissue-specific gene expression (Cai et al., Reference Cai, Wei, Li, Xu and Wang2007). In Arabidopsis, phytochrome-interacting factor 7, which specifically bound to box V with the G-box sequence of the DREB1C promoter, acted as a transcriptional repressor for DREB1C expression. This negative regulation may be important for avoiding plant growth retardation (Kidokoro et al., Reference Kidokoro, Maruyama, Nakashima, Imura, Narusaka, Shinwari, Osakabe, Fujita, Mizoi, Shinozaki and Yamaguchi-Shinozaki2009). In tobacco, a cis-acting region was identified by sequential and internal deletions of the NsCBTS-2a promoter, and its interaction with trans-regulators required for the expression of the CBTS genes restricted to the secretory cell of the glandular trichomes (Ennajdaoui et al., Reference Ennajdaoui, Vachon, Giacalone, Besse, Sallaoud, Herzog and Tissier2010).
In our research, we identified OsERF3 using a yeast one-hybrid screen, in which the bait sequence was the LRK6 promoter DSLP2 region, which is significantly different in 93-11 and Nipponbare varieties. The DSLP2 of var. 93-11 has the whole bait sequence; var. Nipponbare has none. We showed that the cis-element of DSLP2 interacts with the OsERF3 protein, which is characterized as a repressor (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000; Ohta et al., Reference Ohta, Matsui, Hiratsu, Shinshi and Ohme-Takagi2001; Tournier et al., Reference Tournier, Sanchez-Ballesta, Jones, Pesquet, Regad, Latche, Pech and Bouzayen2003).
Phylogenetic tree analysis showed that OsERF3 is a Class II ERF protein, having a conserved EAR motif in its C-terminal region (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000). The domain containing the EAR motif functions as a repression domain, and mediates Class II ERF protein repression activity (Ohta et al., Reference Ohta, Matsui, Hiratsu, Shinshi and Ohme-Takagi2001). Class II ERF repressors down-regulate the transactivation activity of other ERFs (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000), and suppress the activation activity of other ERF proteins when co-expressed (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000; Ohta et al., Reference Ohta, Matsui, Hiratsu, Shinshi and Ohme-Takagi2001). In ethephon treatment experiments, the expression level of LRK6 decreased as the expression of OsERF3 increased after 3 h of treatment in 93-11. This effect was not observed in Nipponbare. Therefore, we suggest that OsERF3 inhibits the expression of LRK6 in 93-11, but has no effect on the expression of LRK6 in Nipponbare, resulting in higher expression of LRK6 in Nipponbare compared with 93-11. This result is consistent with previous studies of the differential expression of LRK6 in Nipponbare and in 93-11 (He et al., Reference He, Luo, Tian, Li, Zhu, Su, Qian, Fu, Wang, Sun and Yang2006), and could also explain the observed differential allelic expression in the Nipponbare/93-11 hybrid. The binding of OsERF3, which is an ethylene-response factor, to the promoter of LRK6, a yield-related gene, might serve to connect plant development with hormones and environmental factors.
The OsERF3 protein may have more than one binding motif
The highly conserved ERF domain is responsible for DNA-binding activity (Cao et al., Reference Cao, Song, Goodman and Zheng2006). ERFs bind the GCC-box (GCCGCC) element present in the promoters of many pathogen-resistant genes (Ohme-Takagi & Shinshi, Reference Ohme-Takagi and Shinshi1995; Solano et al., Reference Solano, Stepanova, Chao and Ecker1998; Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000; Ohme-Takagi et al., Reference Ohme-Takagi, Suzuki and Shinshi2000; Gu et al., Reference Gu, Wildermuth, Chakravarthy, Loh, Yang, He, Han and Martin2002). ERF proteins can also affect gene expression of non-GCC-box-containing genes, either by regulating the expression of other transcription factors, or by interacting physically with other transcription factors involved in growth and development (Buttner & Singh, Reference Buttner and Singh1997; Chakravarthy et al., Reference Chakravarthy, Tuori, D'Ascenzo, Fobert, Despres and Martin2003). In Arabidopsis, AtERF1, AtERF2 and AtERF5 appear to be the most sensitive to single-nucleotide substitutions within their GCC-box sequences, while AtERF3 and AtERF4 appear to be more flexible with respect to their target sequence preferences (Fujimoto et al., Reference Fujimoto, Ohta, Usui, Shinshi and Ohme-Takagi2000). The tobacco ERF protein Tsi1, Arabidopsis CBF1 and tomato JERF1 can bind to both the GCC and the CRT/DRE sequences (Park et al., Reference Park, Park, Lee, Ham, Shin and Paek2001; Hao et al., Reference Hao, Yamasaki, Sarai and Ohme-Takagi2002; Zhang et al., Reference Zhang, Huang, Xie, Chen, Tian, Zhang, Zhang, Lu, Huang and Huang2004). The tomato ERF protein, Pti4, appears to directly regulate gene expression by binding to the GCC box, and possibly to a non-GCC box element, and to indirectly regulate gene expression by either activating the expression of transcription factor genes or by interacting physically with other transcription factors (Chakravarthy et al., Reference Chakravarthy, Tuori, D'Ascenzo, Fobert, Despres and Martin2003). Koyama et al. (Reference Koyama, Okada, Kitajima, Ohme-Takagi, Shinshi and Sato2003) showed that tobacco ERF3 can interact with the B8 protein, but that its ERF or EAR domain alone is insufficient for this interaction; another domain in the ERF protein might be responsible for the interaction. In our research, the promoter of LRK6 has no GCC-box, but can bind an ERF protein via the newly identified motif. In summary, there may be more than one binding motif in ERF proteins. Our results show that OsERF3 can bind both the GCC-box and a new motif, (5′-TAA(A)GT-3′), in vivo. The binding of OsERF3 with the GCC-box might be involved in resistance to biotic and abiotic stresses, and have an important role in rice growth and development. However, this hypothesis remains to be tested.
OsPDCD5 is an effective reporter gene for promoter analysis in rice
To gain an insight into differential allele expression, we studied one member of the LRK gene family, LRK6, which is expressed at much higher levels in Nipponbare rice than in 93-11. Our allele-specific expression data indicated that the allelic expression ratios in hybrids were approximately maintained compared to their parents. Generally, genes with strict cis-regulation have the same bias of expression of the two alleles in both a hybrid and its parents (Kiekens et al., Reference Kiekens, Vercauteren, Moerkerke, Goetghebeur, Van Den Daele, Sterken, Kuiper, Van Eeuwijk and Vuylsteke2006; Zhuang & Adams, Reference Zhuang and Adams2007). We therefore suggest that the majority of variation in LRK6 expression between 93-11 and Nipponbare may be due to cis-regulation. We used 5′ deletions of the LRK6 promoter to identify the candidate cis-element of the promoter, using OsPDCD5 as a reporter gene. OsPDCD5 is highly homologous to human PDCD5, and is up-regulated by low temperature and NaCl treatment (Mi et al., Reference Mi, Wei, Huang, Du, Qian, Li, Shen and Yang2004). Overexpression of OsPDCD5 can induce cell death in transgenic rice plants. Its activity starts in the S2–S3 stage and continues until the complete death of the plant. Other morphological changes include precocious induction of leaf yellowing, early leaf senescence, growth inhibition and early death (Attia et al., Reference Attia, Li, Wei, He, Su and Yang2005). Su et al. (Reference Su, Wu, Wei, Li, He, Attia, Qian and Yang2006) showed that OsPDCD5 is up-regulated during leaf and root senescence. Thus, OsPDCD5 is becoming the reporter gene of choice for 5′ deletion promoter analysis in rice. The LRK6 promoter deletion experiments identified a region, termed DSLP2, which contained a potential cis-element.
This work was supported by the China ‘973’ Foundation (Grant No. 2007CB109002).








