INTRODUCTION
Few if any recent developments in Quaternary science have raised such levels of both interest and contention as ancient DNA (aDNA). Impressive advances have taken place, particularly in the study of mammalian paleoecology, where there have been insights into genetics, lineage development, and extinction (e.g., Shapiro et al., Reference Shapiro, Drummond, Rambaut, Wilson, Matheus, Sher, Pybus, Gilbert, Barnes, Binladen, Willerslev, Hansen, Baryshnikov, Burns, Davydov, Driver, Froese, Harington, Keddie, Kosintsev, Kunz, Martin, Stephenson, Storer, Tedford, Zimov and Cooper2004; Heintzman et al., Reference Heintzman, Froese, Ives, Soares, Zazula, Letts, Andrews, Driver, Hall, Hare, Jass, MacKay, Southon, Stiller, Woywitka, Suchard and Shapiro2016) and also in the understanding of floristic change (e.g., Willerslev et al., Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014). In this forum, I reflect on several decades of research that uses ancient DNA retrieved from sedimentary environments (sedaDNA): ancient DNA deposited in sedimentary environments and subsequently retrieved from deposits such as loess, paleosols, and lacustrine sediments. It is particularly useful in addressing a range of key questions about environmental change. The focus has been on plants to date, but the approach can address many other organismal groups.
Previous sedaDNA studies received some harsh criticism (e.g., Birks and Birks, Reference Birks and Birks2016). The trajectory of ancient DNA publications by decade from the 1980s (Fig. 1) shows a classic evolution. It starts as a topic that is novel and intriguing, then proceeds to the phase called by Jackson (Reference Jackson2012) and Birks and Birks (Reference Birks and Birks2016) the “reification” of a new method, when its contribution is exaggerated and its shortcomings ignored or underplayed. Subsequently, we reach a more rigorous phase of critique, in which the community in general asks whether key issues have been checked and understood, and how various unusual or even inexplicable results are being dealt with. Today, we have come to a place where sedaDNA studies are recognised as a powerful new method in the Quaternary paleoecology repertoire, but with a unique set of problems and pitfalls that require further work to address fully.
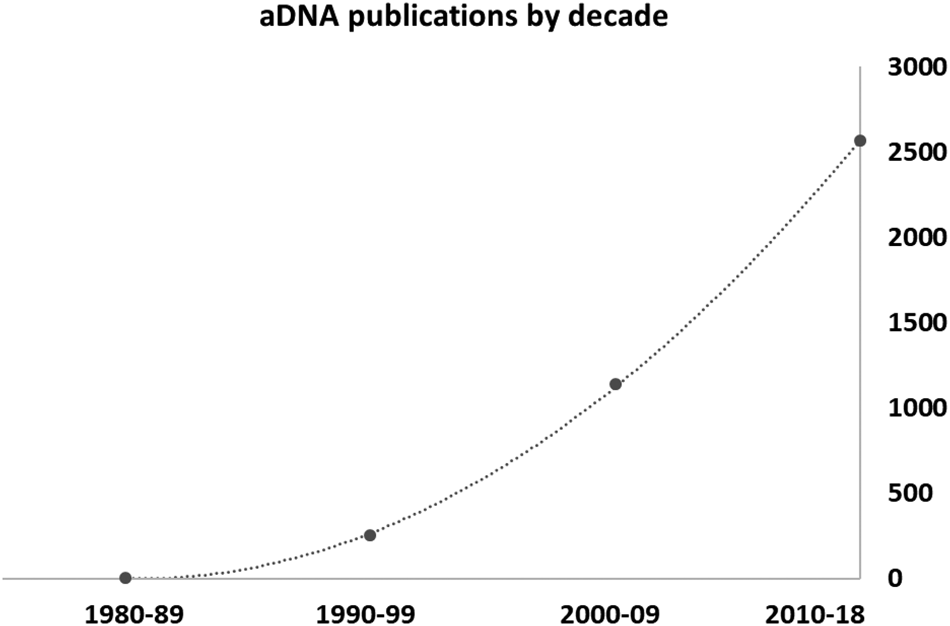
Figure 1. Approximate number of published articles on ancient DNA through time, based on the Web of Knowledge search engine.
The contentious early phases of the field originated in no small part from the lack of reciprocal knowledge among Quaternary paleoecologists on the one hand and molecular biologists on the other. With more and more collaboration occurring across this considerable disciplinary divide, sedaDNA studies are now contributing fully and usefully to Quaternary paleoecology.
Here I attempt a simple overview of the approach, and in doing so address criticisms levelled at it by some traditional paleoecologists with examples of recent studies. As I came to this area from a Quaternary perspective myself, it is clear to me that the fields need to work in partnership, and there is much to be learned on both sides (see also Parducci, Reference Parducci2019). By using the forum, I invite further input from both Quaternary and molecular scientists to update each other on key issues, so that we can advance collaborations even further.
HOW sedaDNA IS STUDIED
Several authors have reviewed methods for environmental and ancient DNA analysis and issues associated with these methods (Pedersen et al., Reference Pedersen, Overballe-Petersen, Ermini, Sarkissian, Haile, Hellstrom, Spens, Thomsen, Bohmann, Cappellini, Schnell, Wales, Caroe, Campos, Schmidt, Gilbert, Hansen, Orlando and Willerslev2015, Taberlet et al., Reference Taberlet, Bonin, Zinger and Coissac2018). Here I provide a brief overview.
Retrieving DNA from sediments
DNA may reside partly in fossil remains such as pieces of plant material but also as extra-cellular DNA in soil solution or bound to mineral particles. The most promising sediments are cold or frozen, anaerobic, around neutral pH and/or dry (as in cave deposits). Clay-silt particles bind to DNA fragments and tend to reduce enzymatic activity in the process, though recovery of sedaDNA may also be successful from organic-rich sediments. Exactly why some sediments yield good results and others not is still under investigation—for example, a recent study has demonstrated that mineral composition and its relation to the charged nature of the sediment appears to have an important effect on DNA retrieval (Freeman et al., ND), underlining the need for a more detailed understanding of sedaDNA taphonomy (see below).
The environmental temperature influences preservation to some extent. Warmer environments are expected to lead to shorter preservation times; in lake sediment, for example, preservation appears to be limited to a few millennia in warm climate (Epp et al., Reference Epp, Stoof, Trauth and Tiedemann2010; Boessenkool et al., Reference Boessenkool, McGlynn, Epp, Taylor, Pimentel, Gizaw, Nemomissa, Brochmann and Popp2014), compared with 104–105 yearsin cold climates. Fluctuating environmental/storage temperatures are also adverse indicators for preservation. As the degree of DNA degradation and/or fragmentation accumulates over time (with rates differing with environmental and storage conditions), the DNA sequences that remain are often short and fragmented.
DNA is usually extracted from sediments in a dedicated clean lab but can also be captured in the field in a sterile buffer solution, and then transferred to the lab (see Taberlet et al., Reference Taberlet, Bonin, Zinger and Coissac2018, p 36). After one of several processing pathways, captured fragments are characterized via sequencing the base pairs of the DNA strand. With the availability of high-throughput sequencing, multiple samples containing a range of molecular taxononmic units (MOTUs) can be analysed simultaneously. MOTUs are specific sequences matching information in available look-up databases that could refer to a single species or be shared at the sub-genus, genus, or, in a few cases, family level.
Extracts of DNA taken from sediments are subject to further treatment before sequences can be identified. Most paleoecological sedaDNA studies to date use metabarcoding, in which specific sequences are aligned with regional catalogues (sequence databases), usually with a focus on a particular oganismal group. Another approach is “shotgun” sequencing, in which all obtained sequences (subject to a practical limit) are compared with a database containing whole genome or partial genome sequences. Each method has its share of imperfections, but each has also yielded exciting results over the past decade.
Shotgun sequencing
Shotgun sequencing has the potential to look for a multitude of taxa from different organismal groups. It is less subject to bias introduced by laboratory processing than metabarcoding (see below), but it generates huge challenges at the stage of bioinformatic analysis. The sheer number of possibilities, combined with a highly incomplete global register of genomic information, mean that many taxa go unrecognised. The accidental sharing of a retrieved sequence by one or more organisms that have no link to the system under study can lead to eyebrow-raising results; these are, ideally, reported for full disclosure but then dismissed via a biogeographic/paleoecological argument. Usefully, the ends of older sequences retrieved using a shotgun approach will show deamination damage (tell-tale runs of unlikely bases), which can confirm whether a sequence or set of sequences is relatively ancient and not modern contamination.
The shotgun approach in paleoecology is exemplified by a study by Pedersen et al. (Reference Pedersen, Ruter, Schweger, Friebe, Staff, Kjeldsen, Mendoza, Beaudoin, Zutter, Larsen, Potter, Nielsenlo, Rainville, Orlando, Meltzer, Kjaer and Willerslev2016) on the western North American ice-free corridor. Using a multi-proxy approach that included sedaDNA, they assessed whether environmental conditions could have allowed human access (via the corridor) prior to or coincident with the first evidence of occupation of regions south of the ice sheet (the data suggest not). This study to some extent suffered from an almost overwhelming signal of many taxa not relevant to the question (e.g. bacterial sequences) that is a common feature of shotgun sequencing. This feature can be circumvented, however, by targeted capture of sequences via specific molecular probes that pick out the sequences of taxa that are of ecological relevance (Taberlet et al., Reference Taberlet, Bonin, Zinger and Coissac2018).
Metabarcoding
The “barcode of life” represents a molecular approach to contemporary taxonomy and identification (Ratnasingham and Herbert, Reference Ratnasingham and Hebert2007). In a similar vein, paleoecologists identify ancient DNA sequences from Quaternary sediments by matching them against short sequences of DNA taken from modern reference organisms. While the majority of studies focus on plants, it is also possible to study fungi, bacteria, mammals and other vertebrates (see below).
Taberlet et al. (Reference Taberlet, Coissac, Pompanon, Gielly, Miquel, Valentini, Vermat, Corthier, Brochmann and Willerslev2007; Reference Taberlet, Bonin, Zinger and Coissac2018) provide detailed information on the metabarcoding technique for plants. The approach uses a short (~10–150 base pair) sequence of DNA (metabarcode) from the plant chloroplast or nuclear genome that is highly variable at the genus and species level as the basis for identification. Retrieved sequences are matched against a regional catalogue of metabarcodes. Bearing in mind that sequences longer than ~100–150 base pairs seldom survive long in extracellular situations due to the action of hydrolyzing enzymes, this restricts the useful areas of the plant genome to those showing high variability in a relatively short sequence (Taberlet et al., Reference Taberlet, Coissac, Pompanon, Gielly, Miquel, Valentini, Vermat, Corthier, Brochmann and Willerslev2007). The use of high-throughput (next-generation) sequencing allows multiple unidentified sequences derived from many samples to be analysed together, generating huge numbers of sequence reads. While complete laboratory and bioinformatic analyses of a set of samples usually takes several weeks or months, the amount of information gained is highly time-effective, though still costly, compared with a more conventional approach such as palynology.
Polymerase chain reaction (PCR) metabarcoding uses primer pairs to capture certain key sequences that can differentiate taxa. As the relevant sequences are rare among all those in the extraction, once captured their numbers are boosted via adding DNA template and polymerase enzymes and multiplying-up the targeted sequences. The total amplified sequence count is likely to reflect the original abundance of different DNA sequences in the sample, but it can be unpredictably biased in either direction (too many or two few), particularly for rare MOTUs (Fig. 2).
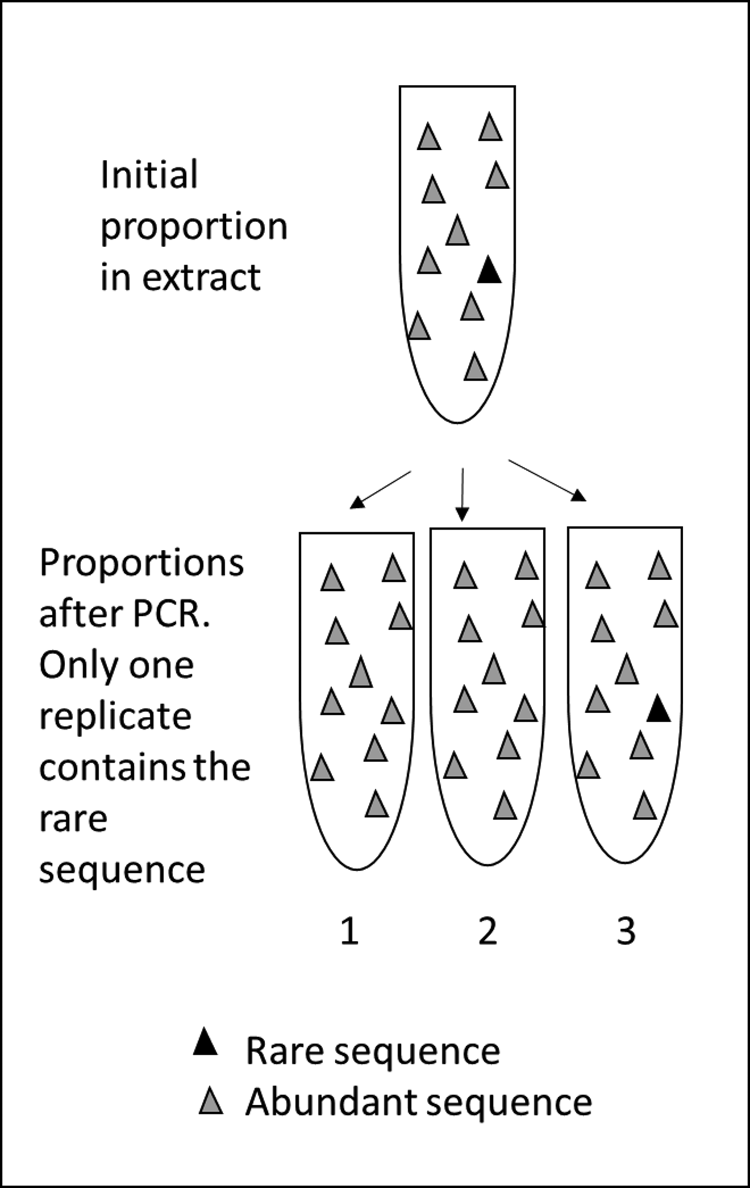
Figure 2. PCR metabarcoding can introduce bias during laboratory processing that is more significant than that affecting other biological proxies. Upper: a “true” proportion of sequences in an extraction prior to PCR. Lower: after PCR, variable capture of sequences by primers means a rare sequence may be missed and never amplified, while after several cycles of amplification, a dominant sequence may swamp other sequences. In this example, the rare sequence is captured in only one of three replicates (see Ficetola et al., Reference Ficetola, Pansu, Bonin, Coissac, Giguet-Covex, De Barba, Gielly, Lopes, Boyer, Pompanon, Raye and Taberlet2015).
PCRs can be repeated for each sample of extracted sedaDNA. Independent amplifications (replicates) are often repeated as many as eight or 12 times. This approach makes rare sequences more likely to be identified than if only one replicate were used, since they are likely to be missed in a single PCR but should be expected in one or more of the repeat PCRs. This is in contrast to taxa with abundant biomass, which tend to appear in all repeats (see, for example, Alsos et al., Reference Alsos, Coissac, Edwards, Foreid, Gielly, Sjogren, Taberlet and Yoccoz2015; Clarke et al., Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019). This modification makes the estimation of relative abundance of MOTUs less subject to random bias than the use of only one PCR per sample (Figs. 2 and 3).
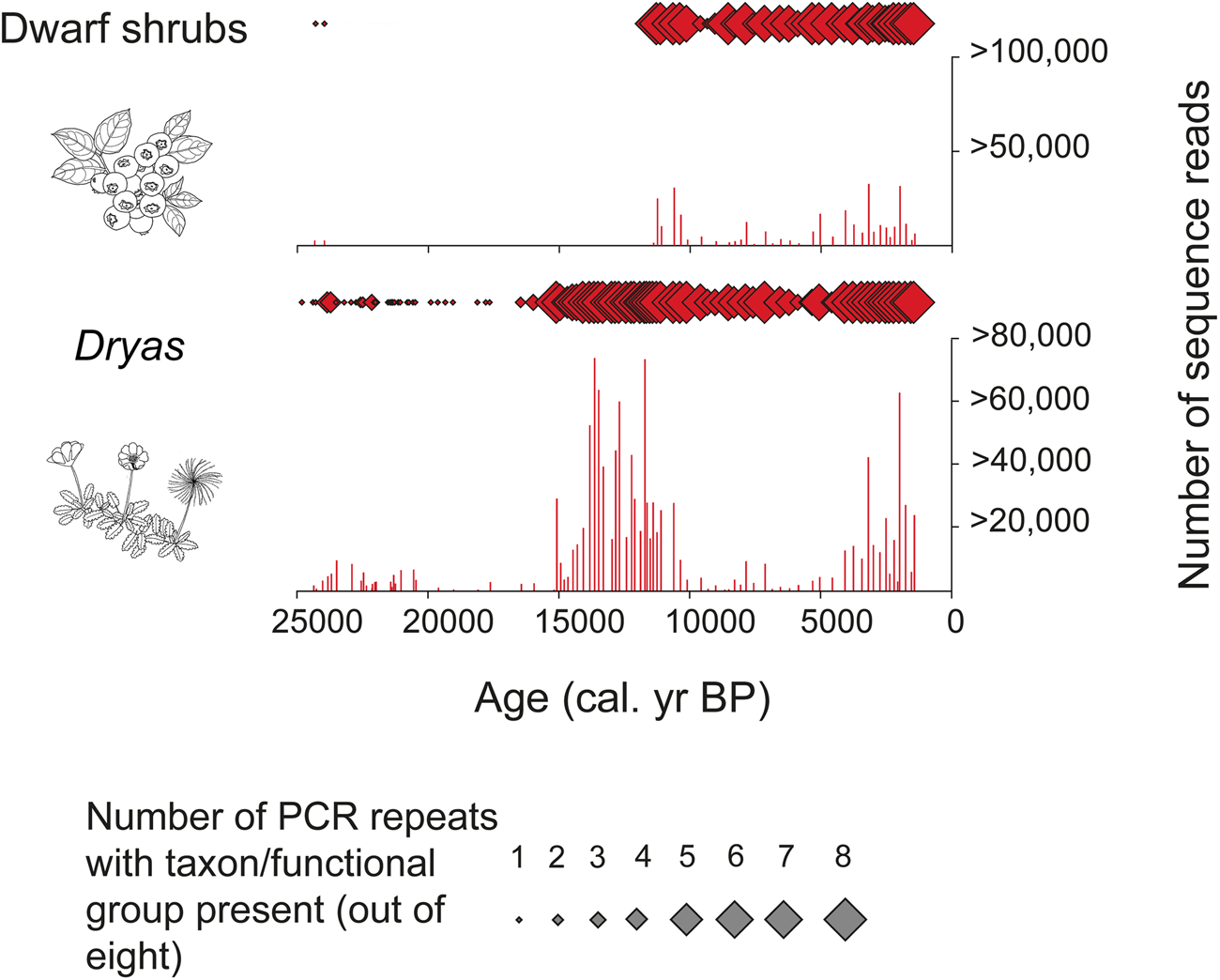
Figure 3. (color online) Numerical abundance of sequence reads (histograms) and proportion of replicates (diamonds) for the functional group “dwarf shrubs” and the genus Dryas through time at Bolshoi Shchuchye Lake, Russia (after Clarke et al., Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019)
Further variation in recovery patterns of sequences relates to the type of polymerase enzyme used (Heintzman et al., Reference Heintzman, Froese, Ives, Soares, Zazula, Letts, Andrews, Driver, Hall, Hare, Jass, MacKay, Southon, Stiller, Woywitka, Suchard and Shapiro2018) and fine details of the PCR process, which are learned and applied in individual labs through experience. These complications increase uncertainty around the estimation of properties such as species richness and may preclude detailed conclusions about relative abundances.
Applied first to modern environmental questions such as animal diet analysis, metabarcoding has subsequently been used in paleoecology to record floristic composition of past vegetation communities, and much more. For example, an early study on DNA retrieved from Quaternary sediments by Willerslev et al. (Reference Willerslev, Hansen, Binladen, Brand, Gilbert, Shapiro, Bunce, Wiuf, Gilichinsky and Cooper2003) demonstrated that small fragments of DNA retrieved from frozen sediment in Siberia contain information on major plant groupings present over late-Quaternary time. While this information did not provide much paleoecological insight, its main message was to demonstrate that ancient, extracellular DNA was preserved and that it could be identified and attributed to taxonomic groups. A decade later the technique had advanced to such a degree that for samples from late-Quaternary sediments across Siberia, Willerslev et al. (Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014) demonstrated the presence of numerous MOTUS. Sites largely comprised frozen yedoma sections (loess-derived deposits; see Kanevskiy et al., Reference Kanevskiy, Shur, Fortier, Jorgenson and Stephani2011; Grosse et al., Reference Grosse, Robinson, Bryant, Taylor, Harper, DeMasi, Kyker-Snowman, Veremeeva, Schirrmeister and Harden2013). The impressive advance in taxononomic richness and resolution was aided by the development of large (>2000 entries) metabarcoding species databases for northern regions (Sonstebø et al., Reference Sonstebo, Gielly, Brysting, Elven, Edwards, Haile, Willerslev, Coissac, Rioux, Sannier, Taberlet and Brochmann2010, Willerslev et al., Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014). A major finding was that, according to the numbers of DNA reads for different plant functional groups, the northern Siberian megafauna may have had access to food sources that comprised forbs as much as they did grasses (by biomass). The extent to which past taxon abundances or biomass might be reflected by the number of occurrences of a particular sequence in a given sample is uncertain and requires further work, however, and thus the quantitative composition of the Pleistocene vegetation remains somewhat speculative.
TAXONOMY AND TAPHONOMY
Critiques of sedaDNA studies mention the limited attainable level of taxonomic resolution (compared with, say, pollen and plant macrofossils), the production of biogeographically incongruous lists of taxa, and a lack of taphonomic understanding (see Birks and Birks, Reference Birks and Birks2016). There were indeed several problems with early studies: they did not use the most complete molecular taxonomy available, and the DNA was then compared with an arbitrary pollen dataset, which also was likely to be taxonomically depauperate (e.g., Pedersen et al., Reference Pedersen, Ginolhac, Orlando, Olsen, Andersen, Holm, Funder, Willerslev and Kjaeer2013). Some spectacular mixtures of possible and improbable taxa were reported from assemblages, without clear warnings about the vagaries of shotgun sequencing (Smith et al., Reference Smith, Momber, Bates, Garwood, Fitch, Pallen, Gaffney and Allaby2015). That some publications tended to focus on how sedaDNA findings compared with, for example, conventional pollen analysis, with little attention to taphonomy or the broader Quaternary context, shows that in some cases data were generated faster than good communication was developed between disciplines. Studies over the past decade reflect greater clarity regarding their physical setting and interpretational constraints; for example, they are placed in clear stratigraphic contexts (e.g., Jorgensen et al., Reference Jorgensen, Haile, Moller, Andreev, Boessenkool, Rasmussen, Kienast, Coissac, Taberlet, Brochmann, Bigelow, Andersen, Orlando, Gilbert and Willerslev2012; Alsos et al., Reference Alsos, Coissac, Edwards, Foreid, Gielly, Sjogren, Taberlet and Yoccoz2015), and several have now explicitly addressed issues of modern representation, i.e., the calibration of modern DNA against modern vegetation (see below). These studies provide an important foundation for ongoing work, though there is much more still to be learned.
Several recent studies investigate the match between near-modern DNA and ecological observations. Yoccoz et al. (Reference Yoccoz, Brathen, Gielly, Haile, Edwards, Goslar, von Stedingk, Brysting, Coissac, Pompanon, Sonstebo, Miquel, Valentini, de Bello, Chave, Thuiller, Wincker, Cruaud, Gavory, Rasmussen, Gilbert, Orlando, Brochmann, Willerslev and Taberlet2012) studied soil DNA in relation to extant plant communities across a landscape in north Norway and found the different plant communities to be reflected in modern soil DNA assemblages. Their data also suggested differential scaling between measured biomass and amount of DNA retrieved for different plant functional types. For example, forb DNA was recovered in a greater proportion to measured forb biomass, whereas the DNA of dwarf shrubs appeared under-represented in relation to biomass. In a study of the relationship of DNA from soil with vegetation on Svalbard, Edwards et al. (Reference Edwards, Alsos, Yoccoz, Coissac, Goslar, Gielly, Haile, Langdon, Tribsch, Binney, von Stedingk and Taberlet2018) showed that 98% of identified DNA taxa were recorded in exhaustive vegetation surveys carried out over a 4.0-m radius around each sample location. Further, 50% of extant plant taxa found within 1.0 m of the soil sampling point were represented in DNA. Thus the DNA-vegetation relationship showed high floristic fidelity, but under-representation of full species richness. Furthermore, identified DNA taxa were most likely to be detected if they occurred within 0.5 m of the sampling point. Very few taxa found in the DNA were located at a greater distance (>1.0 m) from the sampling point (Fig. 4). Even where different taxa grew on slopes above the sampling point, those taxa did not appear in the samples, suggesting negligible local downslope transport.
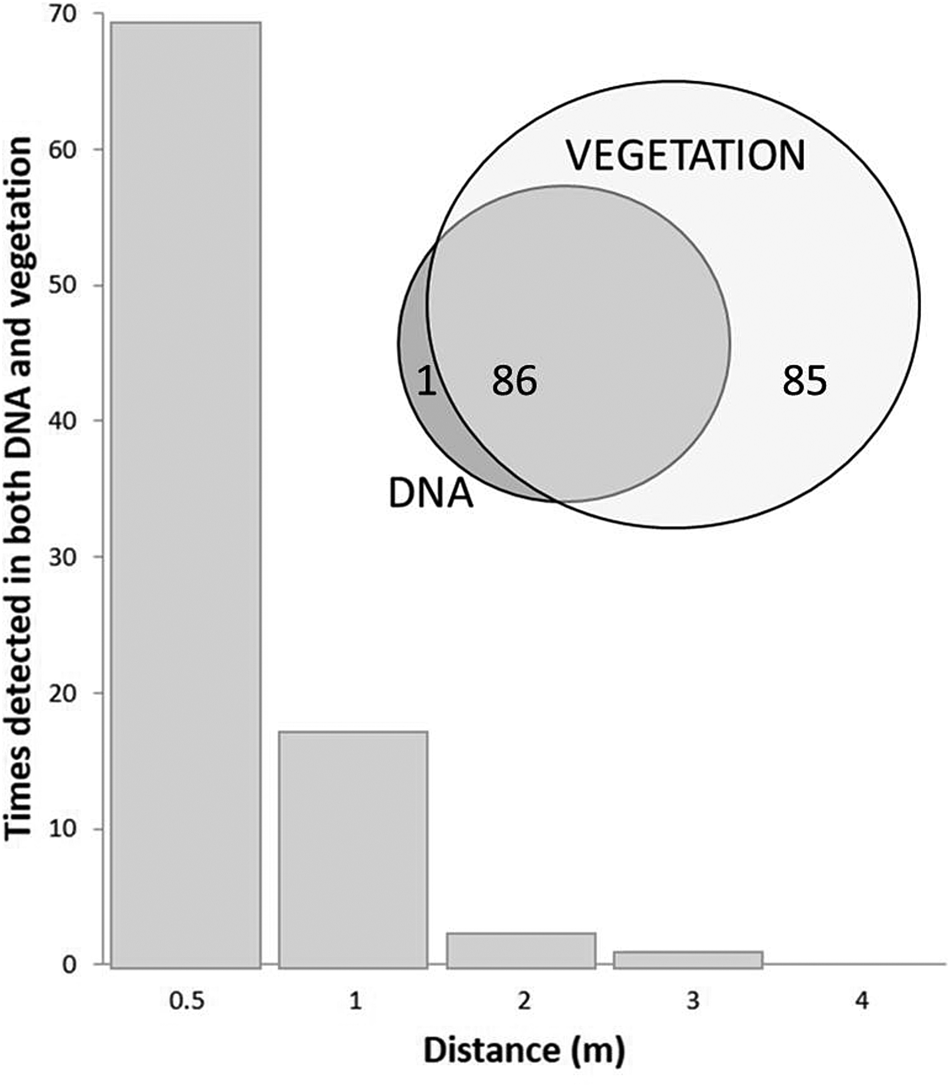
Figure 4. Upper: Svalbard tundra soil samples vs vegetation—all but one recovered MOTU matched observed taxa in vegetation, but only 50% of vegetation taxa were identified in the DNA. Lower: Taxa detected in both vegetation and soil DNA at sampling points. Nearly all such occurrences were found within 0.5 m of the sample point (see Edwards et al., Reference Edwards, Alsos, Yoccoz, Coissac, Goslar, Gielly, Haile, Langdon, Tribsch, Binney, von Stedingk and Taberlet2018).
These findings indicate a local signal should be expected from soil/paleosols, at least in the northern high latitudes. Hence, the results from yedoma/paleosols (as used in Willerselv et al., Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014; Zimmerman et al., Reference Zimmermann, Raschke, Epp, Stoof-Leichsenring, Schirrmeister, Schwamborn and Herzschuh2017; and Zobel et al., Reference Zobel, Davison, Edwards, Brochmann, Coissac, Taberlet, Willerslev and Moora2018) almost certainly represent a highly restricted sampling of the palaeoenvironment. It may also explain the fact that only 100–200 MOTUs, of over 2000 possibilities in the databases, appear in the Willerslev et al. (Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014) dataset, as so little space on the ground was actually sampled.
First steps have been made towards developing a taphonomic theory of sedaDNA. Conceptual taphonomic models are provided by Pedersen et al. (Reference Pedersen, Overballe-Petersen, Ermini, Sarkissian, Haile, Hellstrom, Spens, Thomsen, Bohmann, Cappellini, Schnell, Wales, Caroe, Campos, Schmidt, Gilbert, Hansen, Orlando and Willerslev2015) and Edwards et al. (Reference Edwards, Alsos, Yoccoz, Coissac, Goslar, Gielly, Haile, Langdon, Tribsch, Binney, von Stedingk and Taberlet2018), among others. Much is still to be learned about the pathways leading from the initial death assemblage, when DNA leaches from decomposing cells, to what is eventually preserved in soils and other types of sediment, but the increasing number of studies carried out in different types of sedimentary basins provide a growing basis of empirical evidence. Several recent studies have focused on how DNA enters lake sediments and what the DNA “catchment” is for lakes (e.g., via overland flow, seepage, and groundwater, vs. stream/river input). Further questions relate to the representation of vegetation in DNA from lake-surface sediments, coring contamination, down-core leaching (i.e., is there temporal fidelity?) and whether long-distance pollen contributes to sedaDNA (see, for example, Parducci et al., Reference Parducci, Valiranta, Salonen, Ronkainen, Matetovici, Fontana, Eskola, Sarala and Suyama2015; Sjogren et al., Reference Sjogren, Edwards, Gielly, Langdon, Croudace, Merkel, Fonville and Alsos2017; Alsos et al., Reference Alsos, Lammers, Yoccoz, Jorgensen, Sjogren, Gielly and Edwards2018).
To assess the spatial and temporal reliability of sedaDNA in representing vegetation changes of known extent and date, Sjogren et al. (Reference Sjogren, Edwards, Gielly, Langdon, Croudace, Merkel, Fonville and Alsos2017) studied the recent sedaDNA records of small lakes in Galloway Forest in Scotland, where plantations of non-native conifers were widely established in the 20th century. Soil erosion either occurred prior to any planting or only much later with first harvesting, so reworking of exotic conifer material should not have been a problem. Records from radiometrically dated short cores revealed that the appearance of conifer DNA reflects the time of exotic planting, so it is unlikely there was downward leaching of DNA. A large and ecologically consistent turnover of taxa broadly coincident with planting suggests that the record accurately shows the planting (not the first reproductive activity, i.e., pollen production) of the conifers and a change in the field layer of the forest. The DNA catchment appeared to be the hydrologic catchment, and as there were inflowing streams, the plentiful amount of DNA retrieved may reflect this geographically extended source. There was also high floristic fidelity (all observed taxa occurred in vice-county botanical records), but under-sampling of full floristic diversity.
The importance of inflowing streams and slope-wash is probably reflected in the DNA record from a large lake in the Polar Urals, where the 215-km2 hydrological catchment includes an inflowing river and spans hundreds of meters in elevation (Svendsen et al., Reference Svendsen, Haflidason, Henriksen, Hovland, Mangerud, Nazarov, Lohne, Regnell, Gyllencreutz, Faerseth and Schaefer2019). Over 100 vascular plant MOTUs were retrieved from numerous sequences (Clarke et al., Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019). In complete contrast, a suite of small lakes lacking inflowing streams in north Norway had more variable DNA recovery, and some records were dominated by in-lake taxa. There appeared to be a strong link between the location of terrestrial species at or near the lake margin and their likelihood of occurrence in lake surface-sediments (Alsos et al., Reference Alsos, Lammers, Yoccoz, Jorgensen, Sjogren, Gielly and Edwards2018). In a shotgun approach to a lake-sediment study, Parducci et al. (Reference Parducci, Alsos, Unneberg, Pedersen, Han, Lammers, Salonen, Valiranta, Slotte and Wohlfarth2019) also found that in-lake biomass tended to swamp the terrestrial signal. These observations suggest that the conventions of pollen analysis as regards the best lakes for paleoecological studies (i.e., relatively small, no inlet, often productive) do not necessarily hold for DNA studies, and so the relationship of question to site type may need rethinking.
INTRODUCTION OF CONTAMINATION
Contamination is a problem for many proxy studies. It certainly can occur during collection of sediment samples and cores, but it is now clear from numerous studies in the early days of sedaDNA that draconian anti-contamination measures required by critics and practitioners alike are excessive (see for example, measures taken in Alsos et al., Reference Alsos, Coissac, Edwards, Foreid, Gielly, Sjogren, Taberlet and Yoccoz2015, compared with Clarke et al. Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019). A well-taken and properly treated lake-sediment core, sampled for DNA from undisturbed inner sediments, stands no more chance of contamination than any other proxy. Soil and terrestrial sediment samples, however, do require bleached tools, gloves, and great care while sampling, as here there is more chance of contamination.
There is a greater chance of contamination in the lab. In sedaDNA work, the extremely small sample sizes mean that even reagents can and do contribute contamination (typically, widely occurring foodstuffs). Other possible forms of lab contamination are more subtle and include a still unresolved concern about residue from paper and cardboard products, as Picea and/or Pinus is sometimes filtered out as a potential contaminant (e.g., Alsos et al., Reference Alsos, Sjogren, Brown, Gielly, Foreid Merkell, Paus, Lammers, Edwards, Alm, Leng, Goslar, Langdon, Bakke and van der Bilt2020). For these reasons, it is important always to take measures to identify contamination sources (see below).
Sediment processes, particularly retransportation and reworking, are also of concern. Contention over a sedaDNA study that included sediments in Andøya, Norway (Birks et al., Reference Birks, Giesecke, Hewitt, Tzedakis, Bakke and Birks2012; Parducci et al., Reference Parducci, Edwards, Bennett, Alm, Elverland, Tollefsrud, Jorgensen, Houmark-Nielsen, Larsen, Kjaer, Fontana, Alsos and Willerslev2012a, Reference Parducci, Jorgensen, Tollefsrud, Elverland, Alm, Fontana, Bennett, Haile, Matetovici, Suyama, Edwards, Andersen, Rasmussen, Boessenkool, Coissac, Brochmann, Taberlet, Houmark-Nielsen, Larsen, Orlando, Gilbert, Kjaer, Alsos and Willerslev2012b) lay partly in the possibility that Pinus and Picea DNA occurred because of unrecognized reworking of ancient material into lake sediments. While it is generally assumed that DNA released to aerobic and non-frozen soil will disappear rapidly if not incorporated into frozen/anaerobic deposits, this is not widely demonstrated. Thus, the possibility exists of “ghost” occurrences of DNA due to erosion of old material into a lake. Some evidence that suggests this may not be a major issue comes from the lacustrine sedaDNA record of Clarke et al. (Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019), where boreal forest taxa are present in samples representing the the Holocene thermal optimum but vanish completely from late-Holocene samples, consistent with their absence from the modern vegetation.
Nevertheless, some contamination is almost inevitable in sedaDNA studies. An important element during the processing of sequence data is the filtering out of contaminants, both obvious and subtle. The desired result is to reduce false positives while retaining as many true positives as possible (see Ficetola et al., Reference Ficetola, Pansu, Bonin, Coissac, Giguet-Covex, De Barba, Gielly, Lopes, Boyer, Pompanon, Raye and Taberlet2015). Blank samples at all stages of laboratory processing are used to detect contamination. The total number of reads, or sums of reads across a sample set, or the totals of replicates a sequence is found in, can be used to set reasonable thresholds for positive identification. These criteria are fluid, and much thinking (and some trial and error) is required to set them for a given study (see, for example, Ficetola et al., Reference Ficetola, Pansu, Bonin, Coissac, Giguet-Covex, De Barba, Gielly, Lopes, Boyer, Pompanon, Raye and Taberlet2015; Alsos et al., Reference Alsos, Coissac, Edwards, Foreid, Gielly, Sjogren, Taberlet and Yoccoz2015; Clarke et al., Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019). MOTUs representing taxa that are ecologically and/or biogeographically out of place do pass filters, and thus, as with any other paleoecological study, require assessment by a practioner with good biogeographical and/or molecular knowledge. In speciose genera, a single mistake in a sequence may shift an identification from biogeographically acceptable to clearly wrong, leaving a decision to be made as to how to assign the sequence in question.
DATA ARCHIVING
DNA studies are unusual in the highly complex data that underlie interpretations and conclusions. Making sense of these data and explaining them to audiences not well versed in the details of molecular techniques is important. Furthermore, the coordinated provision of archived data sets that could be used by others in future studies remains an unresolved issue. The vast amounts of bioinformatically processed data stored in genetic databases such as DRYAD (see https://datadryad.org/stash/) are voluminous but may be of little use to many interested in the paleoecological aspects of the study. Quaternary paleoecologists are used to accessing data sorted by site and proxy, accompanied by appropriate metadata such as radiocarbon dates. A given dataset records stratigraphic position, age if available, plus occurrences of identified taxa (see for example, NEOTOMA, https://www.neotomadb.org/). Here, to be widely relevant, the molecular community must adapt to paleoecological norms of data archiving. This is gradually happening, but as yet there is no formal system.
The challenge to those working with sedaDNA is to provide public access to data that are, on the one hand, understandable in paleoecological terms; on the other hand, the procedures and uncertainties in the dataset must be clear. Examples of these procedures and uncertainties are the relationship of reads to replicates; important details of the processing technique; databases consulted to derive MOTU identity; and filtering protocols and contaminant controls. Indeed, given the flexibility inherent in the filtering process, the initial raw data (all MOTU read numbers for all levels and controls) should be available, if required, as well as the filtered dataset used for paleoecological inference. A group being convened by the NEOTOMA team will shortly address such issues, and input from the broader community is welcome (contact the author).
NEW DIRECTIONS FOR QUATERNARY MOLECULAR PALEOECOLOGY
Making use of as many proxies as possible is almost always the best strategy for paleoecological studies. As Birks (Reference Birks and Birks2000) previously reminded us, pollen and macrofossils together are better than either one alone. The same can be said of sedaDNA: using multiple proxies in concert provides some of the strongest inferences. As we understand more about similarities in source area (i.e., DNA more similar to macrofossils than to pollen) and taxonomic resolution (overall pollen and DNA taxonomic resolution is similar in northern regions but differs with respect to taxa; Sonstebø et al., Reference Sonstebo, Gielly, Brysting, Elven, Edwards, Haile, Willerslev, Coissac, Rioux, Sannier, Taberlet and Brochmann2010), we can use the strengths of each method to their best advantage. I conclude this forum with a brief survey of recent studies that illustrate the expanding scope of sedaDNA studies.
Several studies have now demonstrated the power of sedaDNA to evaluate changes through time that feature multiple organismal groups, such as combined stratigraphic records of mammals and plants. Examples include grazing patterns in relation the vegetation cover and erosion in the Alps (Giguet-Covex et al., Reference Giguet-Covex, Pansu, Arnaud, Rey, Griggo, Gielly, Domaizon, Coissac, David, Choler, Poulenard and Taberlet2014), the demise of the Holocene mammoths of St Paul Island (Graham et al., Reference Graham, Belmecheri, Choy, Culleton, Davies, Froese, Heintzman, Hritz, Kapp, Newsom, Rawcliffe, Saulnier-Talbot, Shapiro, Wang, Williams and Wooller2016), and insights into paleo-migration routes (Pedersen et al., Reference Pedersen, Ruter, Schweger, Friebe, Staff, Kjeldsen, Mendoza, Beaudoin, Zutter, Larsen, Potter, Nielsenlo, Rainville, Orlando, Meltzer, Kjaer and Willerslev2016). The addition of sedaDNA analysis to sites associated with archaeological finds should enhance information on features such as the introduction of domestic animals, consequent ecosystem change, the appearance of crop plants, and other aspects of natural resource use. Other molecular proxies from lake-sediment records can reveal human colonization/activity (e.g., coprostanol biomarkers; D'Anjou et al., Reference Anjou, Bradley, Balascio and Finkelstein2012), and potentially this approach can be combined with sedaDNA analysis (see Mackay et al., Reference Mackay, Henderson, van Hardenbroek, Cavers, Crone, Davies, Fonville, Head, Langdon, Matton, McCormick, Murray, Whitehouse and Brown2017).
Critical but cryptic (with regard to proxies) components of ecosystems can now be addressed. For example, Wood et al. (Reference Wood, Diaz, Latorre, Wilmshurst, Burge and Gutierrez2018) use DNA extracted from packrat middens to track pathogen occurrence through changing climatic conditions in the Atacama Desert. Zobel et al. (Reference Zobel, Davison, Edwards, Brochmann, Coissac, Taberlet, Willerslev and Moora2018) report changing mycorrhizal mutualisms through time based on the taxonomically rich Willserslev et al. (Reference Willerslev, Davison, Moora, Zobel, Coissac, Edwards, Lorenzen, Vestergard, Gussarova, Haile, Craine, Gielly, Boessenkool, Epp, Pearman, Cheddadi, Murray, Brathen, Yoccoz, Binney, Cruaud, Wincker, Goslar, Alsos, Bellemain, Brysting, Elven, Sonstebo, Murton, Sher, Rasmussen, Ronn, Mourier, Cooper, Austin, Moller, Froese, Zazula, Pompanon, Rioux, Niderkorn, Tikhonov, Savvinov, Roberts, MacPhee, Gilbert, Kjaer, Orlando, Brochmann and Taberlet2014) dataset by assigning mycorrhizal preferences to the MOTUs. Belle et al. (Reference Belle, Parent, Frossard, Verneaux, Millet, Chronopoulou, Sabatier and Magny2014) record recent changes in lacustrine carbon processing by bacterial methanotrophs. There is an exciting potential to record genetic variation through time in plants. Recently, Lammers et al. (Reference LammersND), using shotgun sequencing, have demonstrated genomic variation at the population level across time in a lacustrine alga, showing the potential for paleoecology to move into paleogenomics.
Although this article has focused on terrestrial paleoecology, it should be noted that sedaDNA is increasingly used in marine settings. For example, Palowska et al. (Reference Pawłowska, Lacka, Kucharska, Pawlowski and Zajaczkowski2020) use diatom DNA in a reconstruction of the history of Stjorfjorden (Svalbard), a critical site for deep-water production, and Giosan et al. (Reference Giosan, Orsi, Coolen, Wuchter, Dunlea, Thirumalai, Munoz, Clift, Donnelly, Galy and Fuller2018) similarly track changes in the planktonic composition of the water column for an Indian Ocean core in a reconstruction of winter monsoon dynamics.
Finally, given concerns about both future climate change and biodiversity loss, sedaDNA can help develop baselines for conservation programmes by contributing to knowledge about past ecosystem state. Clarke et al. (Reference Clarke, Edwards, Gielly, Ehrich, Hughes, Morozova, Haflidason, Mangerud, Svendsen and Alsos2019) use a highly diverse arctic-alpine sedaDNA flora from the Polar Urals site to demonstrate the resilience of mountain plant communities in the face of glacial-deglacial climate change. Wilmshurst et al. (Reference Wilmshurst, Moar, Wood, Bellingham, Findlater, Robinson and Stone2014) combine pollen and sedaDNA records to establish a pre-colonization baseline for ecological restoration on a New Zealand offshore island. Tracking invasive species and documenting their effect is a critical area in conservation biology, and, in an intriguing study on invasion, Ficetola et al. (Reference Ficetola, Poulenard, Sabatier, Messager, Gielly, Leloup, Etienne, Bakke, Malet, Fanget, Storen, Reyss, Taberlet and Arnaud2018) use a 600-year record to document the before-and-after impact of the introduction of rabbits to the sub-Antarctic Kergeulen Islands. As ecology and paleoecology merge to document and understand effects of humans on many aspects of the environment, sedaDNA studies will play an increasingly important role in documenting the extreme and rapidly accelerating changes affecting our planet.
ACKNOWLEDGMENTS
This article resulted from discussions at an international seminar held at the Quaternary Research Center (November 2018); my thanks to Ben Fitzhugh and the QRC for support to attend the meeting. Thanks to Pierre Taberlet, Christian Brochmann, Eske Willerslev and the members of EcoChange working group 2 (EU-FP6: GOCE-2006-036866) for initially inspiring me about ancient DNA. Thanks also to Inger Alsos, Charlotte Clarke and members of the After-Ice team (Norwegian Research Council grant No. 230617/E10) for an interesting and fruitful collaboration on sedaDNA in Norway. Derek Booth, Charlotte Clarke and two reviewers made helpful suggestions about the manuscript, and Charlotte Clarke kindly supplied Figure 3.








