INTRODUCTION
Hepatitis A virus (HAV), a member of the genus Hepatovirus, family Picornaviridae, is a non-enveloped virus resistant to solvent/detergent treatment. HAV is transmitted generally by the faecal–oral route and could be either foodborne or waterborne. However, on rare occasions it can be transmitted parentally through viraemic blood during the incubation period. Outbreaks of HAV infection among haemophilia patients have been reported in Germany, Belgium, Australia and Italy [Reference Mannuccio, Gdovin and Gringeri1–Reference Robertson, Friedberg, Normann, Graff, Flehming and Shouval5].
From October 1998 to March 1999, 57 (4·2%) patients among 1370 registered haemophiliacs were diagnosed with acute hepatitis A. Clotting factors administered to patients were implicated as a source of infection. Detection of HAV in clotting factors can be carried out by molecular methods as HAV does not grow easily in cell culture. Reverse transcription–polymerase chain reaction (RT–PCR) was carried out followed by sequencing to detect HAV in clotting factors and samples from HAV-infected haemophiliacs. There have been reports of clustering of HAV infection detected by molecular methods in haemophiliacs [Reference Mannuccio, Gdovin and Gringeri1–Reference Robertson, Friedberg, Normann, Graff, Flehming and Shouval5]. Therefore, a matched case-control study was carried out in haemophilia patients to investigate the causal relationship between the hepatitis A outbreak and administration of clotting factors.
MATERIALS AND METHODS
Epidemiological investigations
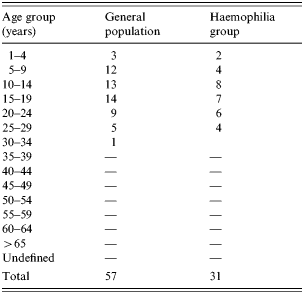
The incidence of hepatitis A was investigated during October 1998 and March 1999. The hospital charts of HAV IgM-positive patients among 1370 haemophiliac patients registered by the Korea Haemophilia Foundation were reviewed. A 1:3 matched case-control study of the patients diagnosed with hepatitis A by HAV IgM test from October 1998 to February 1999 with a record of clotting factor prescriptions was carried out. Of 57 patients with acute HAV, 33 patients whose prescription records were available and received clotting factors were included in this study. An age-matched control group was selected from the registered haemophilia patients who were not diagnosed as having HAV infection by serological tests after October 1998 but who received clotting factors during this period (Table 1).
Table 1. Age distribution of hepatitis A infections in the general population compared with the haemophiliac group
The lots of clotting factors that were prescribed 10–60 days before the hepatitis A infection onset were investigated. Telephone enquiries were carried out to assess other risk factors such as eating uncooked shellfish, travel history, living in closed settings, drinking untreated water and contact with patients with jaundice. Odds ratios (OR) and 95% confidence intervals (CI) for injecting clotting factors and other risk factors were calculated.
Virological investigation
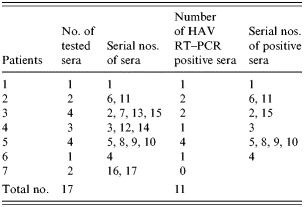
Seventeen sera were tested for the presence of HAV from seven haemophiliac patients and 124 lots of blood clotting factors (both factor VIII and factor IX) which were used during 1998. To prevent any cross-contamination, patients' sera and blood clotting factors were processed separately. Slot and Southern blot hybridization were used to confirm the presence of the HAV genome and the amplified PCR fragments were sequenced.
Total RNA was extracted using Tri-reagent (Molecular Research Centre Inc., Cincinnati, OH, USA) as described in the manufacturer's instructions (Product cat. no. TR-118) from 500 μl of the clotting factors and from 50–100 μl of the patients' sera, followed by isopropanol precipitation at −20°C overnight. Random primers were used for reverse transcription of extracted RNA which was performed at 37°C for 60 min. The enzyme was subsequently inactivated at 95°C for 5 min. For PCR testing, a 247-bp fragment of the VP3/VP1 region of the HAV genome spanning nucleotides 2167–2413 was amplified [Reference Paul, Tada and von der6–Reference Robertson and Khanna8] by cycling at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, repeated 35 times and followed by a 7-min extension at 72°C.
Slot and Southern blot hybridization were performed using a digoxigenin-labelled oligonucleotide probe corresponding to the internal sequence of the VP1 region of HAV (nucleotides 2224–2243). A total of 124 lots of blood clotting factors were tested to confirm the specificity of the PCR amplification and to detect positive samples.
Amplified PCR products were cloned using the TOPO TA cloning kit (Invitrogen Life Technologies, Carlsbad, CA, USA) and sequenced on the ABI Prism 377 automatic sequencer Long Ranger gel. The sequencing reaction was performed in a DNA Thermal Cycler 480 (PerkinElmer, Applied Biosystems, Foster City, CA, USA) by running 25 cycles at 96°C for 30 s, 50°C for 15 s and 69°C for 4 min. The sequences of the PCR products were analysed and compared with other isolates. Nucleotide sequences were analysed using the MegAlign program of Lasergene, DNAStar Inc., Madison, WI, USA. We used the Clustal method to group sequences into clusters by examining the distance between all pairs. The relationship of the sequences is shown in a dendrogram (see Fig.) The length of each pair of branches represents the distance between sequences. Units indicate the numbers of substitution events.

Fig. Nucleotide sequences of the HAV strains detected from clotting factors and patients. The phylogenetic analysis of HAV sequences detected from haemophiliac patients and blood products shows that the sequences of HAV from patient nos. 2, 4, 5 and one blood product (factor VIII, lot no. 1950177) are identical, as are HAV sequences from patient no. 6 and another product (factor IX, lot no. 1510039). The HAV sequence from patient no. 1 is closely related to that from factor IX (lot no. 1510043). MegAlign of the DNAstar aligns multiple sequences by drawing a histogram of consensus strength at the top of each alignment panel using one of two algorithms: the Jotun Hein method or the Clustal method. We used the Clustal method to group sequences into clusters by examining the distance between all pairs. The relationship of the sequences was shown in dendrograms. The length of each pair of branches represents the distance between sequences.
RESULTS
Epidemiological investigations
The incidence of hepatitis A among registered haemophiliac patients was the highest in the 15–19 years age group, followed by the 10–14 and 5–9 years age groups. Examination of regional incidence showed that haemophilia patients in Daejeon had the highest attack rate, although patients were found all over the country except Jeju. Table 2 shows the hepatitis A attack rates among haemophiliacs and non-haemophiliacs by region and Table 3 shows the age distribution.
Table 2. Comparison of attack rates of hepatitis A among haemophiliac and non-haemophiliac patients by region

* Attack rates among males in the general population (%): July 1996 to June 1998.
Table 3. Comparison of attack rates of hepatitis A among haemophiliac and non-haemophiliacs by age group

* Attack rates among males in the general population (%): July 1996 to June 1998.
The hepatitis A incidence rates were 4·5, 3·2 and 0% among patients with haemophilia type A (n=1122), type B (n=189), and other clotting factor disorders (n=59) respectively. Cases of hepatitis A among haemophilia patients were first identified in the first week of October 1998 and peaked in mid October to the final week of October 1998. Another peak (six patients) was detected in December 1998 to January 1999, and four of these patients had type B haemophilia.
The main clinical symptoms of the 50 haemophilia patients whose records were available for review were right upper quadrant abdominal pain, jaundice, fever and gastrointestinal symptoms such as loss of appetite, nausea and vomiting. Their liver function test results also showed high values for serum glutamic-oxaloacetic transaminase (SGOT), serum glutamic-pyruvic transaminase (SGPT), alkaline phosphatase, total bilirubin and direct bilirubin.
From the analysis of risk factors, we have found that one lot (lot no. 1950177) of clotting factor VIII was related significantly to the outbreak of hepatitis A among haemophilia A patients (OR 35, 95% CI 11·3–108·9). An interval of fewer than 10 days between injections was related to a higher incidence of hepatitis A when compared with an interval of more than 10 days (OR 4·2, 95% CI 1·1–15·8). Other factors such as consumption of raw shellfish or other seafood within 2 months of symptom onset, travel or living in a group were not related to a higher incidence of hepatitis A (Table 4). The OR of one lot (lot no. 1950177 of clotting factor VIII was 38·4 (95% CI 8·8–168·2) when the effect of the interval between injections was adjusted.
Table 4. Odds ratio (OR) and 95% confidence intervals (CI) of products among haemophilia A patients

The incidence of hepatitis A and relative risks of the groups who did or did not receive clotting factors from the injection records of 437 individuals was evaluated. A significantly high level of relative risk (46·9, P<0·01) was found in one lot (lot no. 1950177) (Table 5). Statistical analysis was not possible as data were available for only two patients who were injected with factor IX. Although they were injected with lot nos. 1510037, 1510038 and 1510039, the factor IX product of lot no. 1510039 showed the most significant difference in exposure ratios between the case group and the control group.
Table 5. Relative risks of factor VIII among haemophilia patients
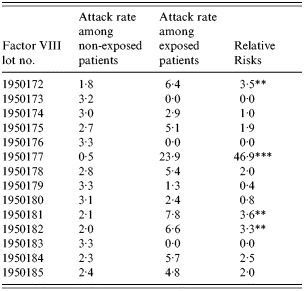
** P < 0·05,
*** P < 0·001.
Virological examinations
Among the 17 sera from seven HAV IgM-positive patients and 124 blood clotting factors (factor VIII and factor IX), HAV RNA was detected in five clotting factors and 11 HAV-positive sera (Tables 6 and 7). HAV RNA was detected in six out of seven patients tested. Five of these six patients and six clotting factors were positive by slot and Southern blot hybridization (Table 6). The analysis of the HAV sequences found in the clotting factors and sera showed that the HAV detected in one of the factors (factor VIII, lot no. 1950177) had 100% sequence homology with HAV found in three patients’ sera (Fig.). This result coincides with the results of the epidemiological analysis in which it was found that those three patients were injected with lot no. 1950177 and this factor was highly related to the HAV infection in haemophilia patients with the high OR (Table 4).
Table 6. Detection of HAV genome from clotting factors
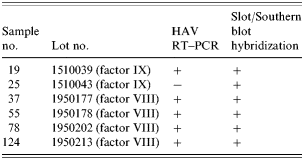
Table 7. Detection of HAV genome from patients
Phylogenetic analysis
The phylogenetic analysis of HAV sequences detected from haemophiliac patients and blood products showed that the HAV sequences from sera 3 (patient no. 4), 10 (patient no. 5), 11 (patient no. 2) and one blood product (factor VIII, lot no. 1950177) were closely related to each other, as were HAV sequences from serum 4 (patient no. 6) and another product (factor IX, lot no. 1510039) (Fig.). The HAV sequence from patient no. 1 was closely related to that of factor IX (lot no. 1510043). However, the HAV sequence from patient no. 3 did not match sequences from any blood products we examined. Sequences from two 1998 Korean isolates (HAV-JUN, HAV-KIM) and a HAV control (HM 175 strain) used as positive controls were not closely related to sequences from patients, although three lots of factors (lot nos. 1950202, 1950178, 1950213) were relatively related closely to two 1998 Korean isolates (HAV-JUN, HAV-KIM).
DISCUSSION
An outbreak of hepatitis A infection was investigated in a group of haemophiliacs in Korea in 1998. A peak in the incidence of hepatitis A was detected from May to June 1998, and the incidence decreased gradually during the latter half of the year. By comparing the incidence of hepatitis A among the general population and haemophilia patients, a significant increase in hepatitis A incidence was detected among the haemophilia patients. The epidemiological study also implicated factor VIII (lot no. 1950177) as a causal agent of this outbreak. Another product (factor IX, lot no. 1510039) was only used in two haemophilia patients, therefore, it was not possible to analyse the relative risk of this lot.
The HAV sequences detected from patient nos. 2, 4, 5 and lot no. 1950177 of factor VIII were identical and closely related to sequences of HAV isolates in the United States. The plasma pool of lot no. 1950177 was imported from the United States. In addition, the HAV sequences from serum 4 (patient no. 6) and clotting factor IX (lot no. 1510039) were identical. Clotting factors used in Korea are produced by the solvent/detergent method to inactivate bloodborne viruses and it has been reported that HAV can survive this procedure [Reference Hollinger and Emerson9].
Although there were reports of HAV strains detected in Korea during 1994–2000, the sequences of these strains could not be compared with the strains from haemophilia patients and from clotting factors because of differences in the amplified regions [Reference Kim, Roh, Jeoung and Kim10–Reference Kwon, Byun and Yeon12]. Comparison of nucleotide sequences from the Korean strains with foreign strains revealed that there were more nucleotide substitutions among the foreign strains.
A causal relationship was found between the injection of blood clotting factors and an outbreak of hepatitis A among haemophilia patients. During 1991–1993, similar cases were reported from other countries including Italy, Germany, Ireland and Belgium where the solvent/detergent method was used to inactivate bloodborne viruses. We recommend that all haemophilia patients in Korea should be vaccinated against HAV following this outbreak.












