INTRODUCTION
The alveolates are defined as a superphylum within a supergroup also containing Stramenopiles and Rhizaria (termed SAR, for Stramenopiles, Alveolata and Rhizaria; Adl et al. Reference Adl, Simpson, Lane, Lukeš, Bass, Bowser, Brown, Burki, Dunthorn, Hampl, Heiss, Hoppenrath, Lara, Le Gall, Lynn, McManus, Mitchell, Mozley-Stanridge, Parfrey, Pawlowski, Rueckert, Shadwick, Schoch, Smirnov and Spiegel2012), and are united by the presence of namesake sub-membranous flattened vesicles termed alveoli. The alveolates include the ciliates, such as Tetrahymena and Paramecium; dinoflagellates, including important pathogens of shellfish; Perkinsus, which represents a branch related to dinoflagellates; the chromerids, such as Chromera and Vitrella, and the closely related colpodellids; and the obligate parasites termed Apicomplexa, including the human pathogens, Toxoplasma and the malaria parasite Plasmodium. Apicomplexans encompass a spectrum of parasitic transmission strategies, such as the transmission via environmentally durable oocysts, including Cryptosporidium, Gregarina and the coccidians Toxoplasma and Eimeria; or via insect vectors, as for the mosquito-borne Plasmodium, and tick-transmitted Babesia and Theileria. Apicomplexans have evolved to specifically target a variety of host cells, such as gut epithelial cells in the instance of Cryptosporidium and Gregarina; gut epithelial recognition followed by disseminative infections having universal host cell invasion and development with regard to Toxoplasma; and life cycle stage-dependent host cell recognition in the insect-transmitted parasites, including pathology-relevant erythrocytic stages with respect to Plasmodium and Babesia, and lymphocytes in the case of Theileria.
Apicomplexans are named for their apical complex which imparts cell polarity for specific interactions with target cells, and provides a conduit for secretions from organelles, such as rhoptries and micronemes. However, this structure is not unique to apicomplexans. For example, the predatory alveolate and close cousin to Apicomplexa, Colpodella, as well as Perkinsus, have highly developed apical complexes and secretion systems (Brugerolle, Reference Brugerolle2002). Apicomplexans might be further defined and unified as having the additional hallmarks of obligate parasitism, and the capacity for gliding motility. This review seeks to illuminate, using manually curated extracellular proteome predictions derived from new whole genome and transcriptome annotations, the evolutionary leap between a hypothetical ‘proto-apicomplexan’, and a last common ancestor within the Apicomplexa.
Ultrastructure studies give a picture of the breadth of apical complex structures from apicomplexans and closely related alveolates such as Perkinsus, Colpodella and the chromerids. These structures and their component secretory organelles loosely range from apical polarity and secretion in Perkinsus (Coss et al. Reference Coss, Robledo and Vasta2001); pseudo-conoid apical complexes, as in Chromera (Moore et al. Reference Moore, Oborník, Janouškovec, Chrudimský, Vancová, Green, Wright, Davies, Bolch, Heimann, Slapeta, Hoegh-Guldberg, Logsdon and Carter2008; Oborník et al. Reference Oborník, Vancová, Lai, Janouškovec, Keeling and Lukeš2011) and Vitrella (Oborník et al. Reference Oborník, Modrý, Lukeš, Cernotíková-Stříbrná, Cihlář, Tesařová, Kotabová, Vancová, Prášil and Lukeš2012); a specialized apical region capable of forming apparent tight junctions, as in Colpodella (Simpson and Patterson, Reference Simpson and Patterson1996; Brugerolle, Reference Brugerolle2002); and the namesake apical regions of apicomplexans conferring apical adhesion and secretion, and mediation of gliding motility, invasion of target cells and tissue disruption (reviewed in Gubbels and Duraisingh, Reference Gubbels and Duraisingh2012). Alveolates have diverse secretory systems at their disposal, organelles whose composition and character differ based upon genera and the life cycle stage. Examples include the broad variety of trichocyst-like secretory organelles of ciliates which release protein and pigment cargos involved in predatory and defence mechanisms (reviewed in Lobban et al. Reference Lobban, Hallam, Mukherjee and Petrich2007; Briguglio and Turkewitz, Reference Briguglio and Turkewitz2014), dense granules having diverse structures and phylogenetic distribution, and apical rhoptries and micronemes (reviewed in Gubbels and Duraisingh, Reference Gubbels and Duraisingh2012). Regulated secretion via calcium fluxes might be a common theme underpinning alveolates ranging from Paramecium to Plasmodium (reviewed in Vayssié et al. Reference Vayssié, Skouri, Sperling and Cohen2000; Plattner et al. Reference Plattner, Sehring, Mohamed, Miranda, De Souza, Billington, Genazzani and Ladenburger2012); in part utilizing conserved plant-like calcium-dependent protein kinases such as described for regulated exocytosis (Lourido et al. Reference Lourido, Shuman, Zhang, Shokat, Hui and Sibley2010) and parasite egress from host cells (McCoy et al. Reference McCoy, Whitehead, van Dooren and Tonkin2012) in Toxoplasma, and processes of Plasmodium (reviewed in Holder et al. Reference Holder, Ridzuan and Green2012). The use of secretory organelles as taxonomic morphological markers is, it might be argued, still in its infancy as a phylogenetic tool; and to our knowledge no cellular localization studies have mapped secretory proteins to their resident organelles in alveolates other than apicomplexans. Indeed, thus far no candidate orthologues for, say, microneme or rhoptry proteins have been identified which are conserved in both pre-apicomplexans and apicomplexans, such as by bioinformatics screens or immunolocalization assays. For Apicomplexa there is a rich, albeit sometimes muddy literature describing constituent proteins of rhoptry, microneme and dense granule organelles. We will not attempt to review that literature here; rather, we will present examples to give an overview comparison of apicomplexan and pre-apicomplexan alveolate extracellular domains and domain architectures in proteins.
Based on ultrastructure and life strategy criteria, in addition to phylogenetic analyses, Colpodella might be considered to be a compelling candidate for the ‘last common ancestor’ of the Apicomplexa (Brugerolle, Reference Brugerolle2002; Leander et al. Reference Leander, Kuvardina, Aleshin, Mylnikov and Keeling2003). This protozoan is capable of specific recognition of prey, such as the kinetoplastid Bodo caudatus; adhering to it and forming a junction via its apical complex; and engulfing the target cell into its gullet-like compartment (Simpson and Patterson, Reference Simpson and Patterson1996; Brugerolle, Reference Brugerolle2002). Colpodella is not parasitic, unlike its cousin, the intracellular pathogen, Perkinsus. The molecular and mechanical details underpinning the invasion strategy of bivalve haemocytes by Perkinsus are not known (Soudant et al. Reference Soudant, Chu and Volety2013) and warrant further study, particularly utilizing the recently released high coverage genome sequence information for this alveolate pathogen. The development of cell invasion or parasitism does not necessarily lie along the evolutionary pathway to the Apicomplexa, as these features have evolved multiple times in the alveolates, including Perkinsus (Soudant et al. Reference Soudant, Chu and Volety2013), and the endoparasite of fish epithelial tissue, the ciliate Ichthyophthirius multifiliis (Coyne et al. Reference Coyne, Hannick, Shanmugam, Hostetler, Brami, Joardar, Johnson, Radune, Singh, Badger, Kumar, Saier, Wang, Cai, Gu, Mather, Vaidya, Wilkes, Rajagopalan, Asai, Pearson, Findly, Dickerson, Wu, Martens, Van de Peer, Roos, Cassidy-Hanley and Clark2011). Within the perkinsids different host targets exist, including the exquisite example of Parvilucifera prorocentri, which invades and develops within dinoflagellates (Hoppenrath and Leander, Reference Hoppenrath and Leander2009). As detailed in the sections following, our descriptions of predicted extracellular proteins suggest that the endosymbiotic chromerids and predatory colpodellids are more akin to Apicomplexa than Perkinsus, in support of phylogenetic analyses (Moore et al. Reference Moore, Oborník, Janouškovec, Chrudimský, Vancová, Green, Wright, Davies, Bolch, Heimann, Slapeta, Hoegh-Guldberg, Logsdon and Carter2008; Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015).
Physical interactions with foreign cells among alveolates include symbiotic or commensal relationships, predation and avoidance thereof, and parasitism. These interactions would be expected to be driving forces in the selection for specific extracellular proteins that underpin recognition, adherence and response to target cells. For example, Plasmodium sporozoites interact with a diverse set of host tissues – firstly salivary gland tissue in the mosquito, and subsequently tropism to hepatocytes in the liver – utilizing receptor-mediated recognition of target cells (reviewed in Sinnis and Coppi, Reference Sinnis and Coppi2007). Alveolate recognition of the environment also includes positive and negative taxis in response to gradients of nutrients, toxins, light and gravity (Eckert, Reference Eckert1972; Fenchel and Finlay, Reference Fenchel and Finlay1984; Francis and Hennessey, Reference Francis and Hennessey1995; Hemmersbach et al. Reference Hemmersbach, Volkmann and Hader1999; Selbach and Kuhlmann, Reference Selbach and Kuhlmann1999; Cadetti et al. Reference Cadetti, Marroni, Marangoni, Kuhlmann, Gioffré and Colombetti2000; reviewed in Echevarria et al. Reference Echevarria, Wolfe, Strom and Taylor2014), as well as avoidance interactions with predators (Knoll et al. Reference Knoll, Haacke-Bell and Plattner1991; Hamel et al. Reference Hamel, Fisch, Combettes, Dupuis-Williams and Barouod2011). A G-protein-coupled receptor having 7-transmembrane domains was characterized in Tetrahymena and shown to be involved in chemoattraction (Lampert et al. Reference Lampert, Coleman and Hennessey2011). Some alveolates, such as in Nassula citrea, have evolved eyespots, and even a primitive lens termed an ocelloid, as in the dinoflagellates, Erythropsidinium and Nematodinium (Gomez, Reference Gomez2008; Hayakawa et al. Reference Hayakawa, Takaku, Hwang, Horiguchi, Suga, Gehring, Ikeo and Gojobori2015; Gavelis et al. Reference Gavelis, Hayakawa, White, Gojobori, Suttle, Keeling and Leander2015). The dinoflagellate Oxyrrhis marina (Slamovits et al. Reference Slamovits, Okamoto, Burri, James and Keeling2011), as well as chromerids and colpodellids, possess amplified gene families of rhodopsin-like proteins, such as exemplified by Cvel_15171.t1 in Chromera, Vbra_19386.t1 in Vitrella and BE-2_cDNA_131008@a107687_15 in Alphamonas edax.
Self–self recognition during fertilization or ciliate conjugation is another physical cell–cell interaction which appears to be mediated by surface proteins, such as the mating type surface protein mtA in Paramecium (Sonneborn, Reference Sonneborn1938; Byrne, Reference Byrne1973; Singh et al. Reference Singh, Saudemont, Guglielmi, Arnaiz, Goût, Prajer, Potekhin, Przybòs, Aubusson-Fleury, Bhullar, Bouhouche, Lhuillier-Akakpo, Tanty, Blugeon, Alberti, Labadie, Aury, Sperling, Duharcourt and Meyer2014). MtA is 1275 amino acids long (e.g. XP_001450586·1 in Paramecium tetraurelia) and has a structure of a N-terminal signal peptide, multiple transmembrane domains at the carboxyl terminus, and predicted extracellular cysteine-rich furin-like domains adjacent the transmembrane region. In P. tetraurelia, mtA appears to have 3 or more possible paralogues of similar architecture, although it is not known if these proteins participate in mating recognition or have other functions. Multi-transmembrane domain proteins with predicted extracellular furin-like domains are found within amplified families in other ciliates, such as Tetrahymena (e.g. TTHERM_01337410) and Oxytricha; but without functional studies it is not possible to speculate if some of these proteins participate in conjugation. Self-recognition proteins have also been described for gamete fertilization in Plasmodium; namely, members of an amplified gene family which encodes glycophosphatidyl inositol (GPI)-linked proteins composed of 6-cys domains (Van Dijk et al. Reference Van Dijk, van Schaijk, Khan, van Dooren, Ramesar, Kaczanowski, van Genert, Kroeze, Stunnenberg, Eling, Sauerwein, Waters and Janse2010) and the HAP2 protein which functions in membrane fusion (Liu et al. Reference Liu, Tewari, Ning, Blagborough, Garbom, Pei, Grishin, Steele, Sinden, Snell and Billker2008). The 6-cys domain proteins are related to the Toxoplasma and Eimeria GPI-linked surface coat SAG proteins, and have been proposed to have originated via lateral transfer of metazoan ephrin proteins (Gerloff et al. Reference Gerloff, Creasey, Maslau and Carter2005; Arredondo et al. Reference Arredondo, Cai, Takayama, MacDonald, Anderson, Aravind, Clore and Miller2012; Reid et al. Reference Reid, Blake, Ansari, Billington, Browne, Bryant, Dunn, Hung, Kawahara, Miranda-Saavedra, Malas, Mourier, Naghra, Nair, Otto, Rawlings, Rivailler, Sanchez-Flores, Sanders, Subramaniam, Tay, Woo, Wu, Barrell, Dear, Doerig, Gruber, Ivens, Parkinson, Rajandream, Shirley, Wan, Berriman, Tomley and Pain2014). Ten members of the family are present in Plasmodium and appear to have roles in multiple stages of the lifecycle, including hepatocyte and intraerythrocytic stages (Ishino et al. Reference Ishino, Chinzei and Yuda2005; Sanders et al. Reference Sanders, Gilson, Cantin, Greenbaum, Nebl, Carucci, McConville, Schofield, Hodder, Yates and Crabb2005; Annoura et al. Reference Annoura, van Schaijk, Ploemen, Sajid, Lin, Vos, Dinmohamed, Inaoka, Rijpma, van Gemert, Chevalley-Maurel, Kiełbasa, Scheltinga, Franke-Fayard, Klop, Hermsen, Kita, Gego, Franetich, Mazier, Hoffman, Janse, Sauerwein and Khan2014).
Surface proteins mediating recognition of the environment might either be anchored within the surface membrane by multiple transmembrane domains, such as in rhodopsins or signalling channels; single transmembrane domains, such as TRAP/MIC2 receptors described in a section below; tethering to membranes by GPI moieties, including ciliate immobilization antigens and the circumsporozoite coat protein of Plasmodium sporozoites; or via interaction with other membrane-anchored proteins, such as the Plasmodium gamete surface protein, P230 (termed Pfs230 in Plasmodium falciparum; Williamson et al. Reference Williamson, Criscio and Kaslow1993). Globular domains within such extracellular proteins might have arisen by vertical inheritance, in many instances followed by lineage-specific divergence such that their origin is obscure; or by lateral transfer (reviewed in Aravind et al. Reference Aravind, Anantharaman, Zhang, de Souza and Iyer2012). The GPI-linked immobilization antigens of ciliates were the first alveolate cell surface proteins to be identified; and the characterization of agglutination by specific immune sera helped to formulate concepts of antigenic diversity, antigenic switching and allelic exclusion (reviewed in Caron and Meyer, Reference Caron and Meyer1989; Beale and Preer, Reference Beale and Preer2008). These themes were of value later in describing the immune pressure driven antigenic diversity and switching in the surface proteins of the kinetoplastid and human pathogen, Trypanosoma brucei, as well as Plasmodium. Despite a wealth of literature, it remains unknown why ciliates devote themselves to great amplification of genes encoding surface coat immobilization antigens (e.g. the family exemplified by P. tetraurelia protein AAA61739·2), and appear to switch expression of these genes. Instances of gene amplification of predicted surface and secreted proteins are frequently repeated in the alveolates, and might be driven by multiple mechanisms. For example, ciliates possess extensive amplifications of genes encoding membrane attack complex perforin (MACPF)-like domains (e.g. protein family exemplified by TTHERM_01380980 in Tetrahymena) which may participate in lytic pore formation, as do the macrophage MACPF proteins of vertebrates, and mediate membrane traversal in apicomplexans such as Toxoplasma and Plasmodium (reviewed in Kafsack and Carruthers, Reference Kafsack and Carruthers2012). It is not known why ciliates require large numbers of MACPF domain encoding genes – perhaps they serve as attack complexes in predation, or in defence from predators – and if the functions and pressures driving gene amplification are conserved or differ for the MACPF gene expansions observed in apicomplexans. Many other examples of extensive amplifications of genes encoding predicted extracellular alveolate proteins are described in the following sections.
ALVEOLATE PHYLOGENY, APICOMPLEXANS AND ‘PROTO-APICOMPLEXANS’
The phylogenetic tree depicted in Fig. 1 gives our current understanding of the relationships of the pre-apicomplexan alveolates with respect to the Apicomplexa, with the chromerids and colpodellids branching at the base of the apicomplexan clade (Janouškovec et al. Reference Janouškovec, Tikhonenkov, Burki, Howe, Kolísko, Mylnikov and Keeling2015; Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015). The placements of specific genera within this general classification of Alveolata is an ongoing work, requiring solutions to many puzzles concerning phylogenetic relationships, such as the affinity of Perkinsus and Colponema with respect to chromerids and dinoflagellates, and the nature of the candidate last-common relatives leading to the Apicomplexa. The chromerids and colpodellids are the most closely related clades to the Apicomplexa, with Gregarina and Cryptosporidium serving as bookends on the apicomplexan side of the transition to obligate parasitism (Templeton et al. Reference Templeton, Enomoto, Chen, Huang, Lancto, Abrahamsen and Zhu2010). These relationships can then be used, coupled with new understandings of ultrastructures and life strategies (e.g. see Okamoto and Keeling, Reference Okamoto and Keeling2014; Portman and Slapeta, Reference Portman and Slapeta2014), as a foundation for examining the transition from a hypothetical ‘proto-apicomplexan’ (indicated by the yellow shaded box in Fig. 1) to the phylum Apicomplexa, using the repertoires of predicted extracellular proteins. As a basis for this review, we have performed extensive and sensitive basic local alignment search tool (BLAST) screens of extracellular proteins in order to compare apicomplexans with other alveolates and to take advantage of new genome sequence information from the ciliates, Ichthyophthirius (Coyne et al. Reference Coyne, Hannick, Shanmugam, Hostetler, Brami, Joardar, Johnson, Radune, Singh, Badger, Kumar, Saier, Wang, Cai, Gu, Mather, Vaidya, Wilkes, Rajagopalan, Asai, Pearson, Findly, Dickerson, Wu, Martens, Van de Peer, Roos, Cassidy-Hanley and Clark2011), Oxytricha (Swart et al. Reference Swart, Bracht, Magrini, Minx, Chen, Zhou, Khurana, Goldman, Nowacki, Schotanus, Jung, Fulton, Ly, McGrath, Haub, Wiggins, Storton, Matese, Parsons, Chang, Bowen, Stover, Jones, Eddy, Herrick, Doak, Wilson, Mardis and Landweber2013) and Stylonychia (Aeschlimann et al. Reference Aeschlimann, Jönnsson, Postberg, Stover, Petera, Lipps, Nowacki and Swart2014); the chromerids, Chromera velia and Vitrella brassicaformis (Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015); Perkinsus marinus; high coverage transcriptome sequence information for several colpodellids (Janouškovec et al. Reference Janouškovec, Tikhonenkov, Burki, Howe, Kolísko, Mylnikov and Keeling2015) and GenBank-deposited genome sequence information for the apicomplexans Gregarina niphandrodes, Cryptosporidium muris, Eimeria tenella and Hammondia. Genome sequence information is also available for Tetrahymena thermophila (Eisen et al. Reference Eisen, Coyne, Wu, Wu, Thiagarajan, Wortman, Badger, Ren, Amedeo, Jones, Tallon, Delcher, Salzberg, Silva, Haas, Majoros, Farzad, Carlton, Smith, Garg, Pearlman, Karrer, Sun, Manning, Elde, Turkewitz, Asai, Wilkes, Wang, Cai, Collins, Stewart, Lee, Wilamowska, Weinberg, Ruzzo, Wloga, Gaertig, Frankel, Tsao, Gorovsky, Keeling, Waller, Patron, Cherry, Stover, Krieger, del Toro, Ryder, Williamson, Barbeau, Hamilton and Orias2006), P. tetraurelia (Aury et al. Reference Aury, Jaillon, Duret, Noel, Jubin, Porcel, Ségurens, Daubin, Anthouard, Aiach, Arnaiz, Billaut, Beisson, Blanc, Bouhouche, Câmara, Duharcourt, Guigo, Gogendeau, Katinka, Keller, Kissmehl, Klotz, Koll, Le Mouël, Lepère, Malinsky, Nowacki, Nowak, Plattner, Poulain, Ruiz, Serrano, Zagulski, Dessen, Bétermier, Weissenbach, Scarpelli, Schächter, Sperling, Meyer, Cohen and Wincker2006), Babesia bovis (Brayton et al. Reference Brayton, Lau, Herndon, Hannick, Kappmeyer, Berens, Bidwell, Brown, Crabtree, Fadrosh, Feldblum, Forberger, Haas, Howell, Khouri, Koo, Mann, Norimine, Paulsen, Radune, Ren, Smith, Suarez, White, Wortman, Knowles, McElwain and Nene2007) and other Babesia species (Cornillot et al. Reference Cornillot, Hadj-Kaddour, Dassouli, Noel, Ranwez, Vacherie, Augagneur, Brès, Duclos, Randazzo, Carcy, Debierre-Grockiego, Delbecq, Moubri-Ménage, Shams-Eldin, Usmani-Brown, Bringaud, Wincker, Vivarès, Schwarz, Schetters, Krause, Gorenflot, Berry, Barbe and Ben Mamoun2012; Jackson et al. Reference Jackson, Otto, Darby, Ramaprasad, Xia, Echaide, Farber, Gahlot, Gamble, Gupta, Gupta, Jackson, Malandrin, Malas, Moussa, Nair, Reid, Sanders, Sharma, Tracey, Quail, Weir, Wastling, Hall, Willadsen, Lingelbach, Shiels, Tait, Berriman, Allred and Pain2014), Eimeria spp. (Heitlinger et al. Reference Heitlinger, Spork, Lucius and Dieterich2014; Reid et al. Reference Reid, Blake, Ansari, Billington, Browne, Bryant, Dunn, Hung, Kawahara, Miranda-Saavedra, Malas, Mourier, Naghra, Nair, Otto, Rawlings, Rivailler, Sanchez-Flores, Sanders, Subramaniam, Tay, Woo, Wu, Barrell, Dear, Doerig, Gruber, Ivens, Parkinson, Rajandream, Shirley, Wan, Berriman, Tomley and Pain2014), Theileria parva and Theileria annulata (Gardner et al. Reference Gardner, Bishop, Shah, de Villiers, Carlton, Hall, Ren, Paulsen, Pain, Berriman, Wilson, Sato, Ralph, Mann, Xiong, Shallom, Weidman, Jiang, Lynn, Weaver, Shoaibi, Domingo, Wasawo, Crabtree, Wortman, Haas, Angiuoli, Creasy, Lu, Suh, Silva, Utterback, Feldblyum, Pertea, Allen, Nierman, Taracha, Salzberg, White, Fitzhugh, Morzaria, Venter, Fraser and Nene2005; Pain et al. Reference Pain, Renauld, Berriman, Murphy, Yeats, Weir, Kerhornou, Aslett, Bishop, Bouchier, Cochet, Coulson, Cronin, de Villiers, Fraser, Fosker, Gardner, Goble, Griffiths-Jones, Harris, Katzer, Larke, Lord, Maser, McKellar, Mooney, Morton, Nene, O'Neil, Price, Quail, Rabbinowitsch, Rawlings, Rutter, Saunders, Seeger, Shah, Squares, Squares, Tivey, Walker, Woodward, Dobbelaere, Langsley, Rajandream, McKeever, Shiels, Tait, Barrell and Hall2005), Cryptosporidium parvum and Cryptosporidium hominis (Abrahamsen et al. Reference Abrahamsen, Templeton, Enomoto, Abrahante, Zhu, Lancto, Deng, Liu, Widmer, Tzipori, Buck, Xu, Bankier, Dear, Konfortov, Spriggs, Iyer, Anantharaman, Aravind and Kapur2004; Xu et al. Reference Xu, Widmer, Wang, Ozaki, Alves, Serrano, Puiu, Manque, Akiyoshi, Mackey, Pearson, Dear, Bankier, Peterson, Abrahamsen, Kapur, Tzipori and Buck2004) and numerous Plasmodium species (available and described at the online database resource, http://www.plasmodb.org).

Fig. 1. Phylogenetic tree of the alveolates, showing relationships with the Apicomplexa (generalized from Templeton et al. Reference Templeton, Enomoto, Chen, Huang, Lancto, Abrahamsen and Zhu2010; Janouškovec et al. Reference Janouškovec, Tikhonenkov, Burki, Howe, Kolísko, Mylnikov and Keeling2015; Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015). The apicomplexan clade is indicated by a grey box. The grouping indicated by the yellow box shows the relationship of the Chromera, Vitrella and the colpodellids; and as a sister clade to the Apicomplexa and thereby putative model ‘proto-apicomplexans’.
GLYCOSYLATION, MUCINS AND SUGAR-BINDING DOMAINS
The fundamental cell–cell interaction of alveolates is probably recognition of carbohydrate residues on target cells mediated by cell surface proteins containing lectin-like domains (Robert et al. Reference Robert, Zubhov, Martin-Cereceda, Novarino and Wootton2006; Wood-Charlson et al. Reference Wood-Charlson, Hollingsworth, Krupp and Weis2006; Wootton et al. Reference Wootton, Zubkov, Jones, Jones, Martel, Thornton and Roberts2007; Martel, Reference Martel2009). Such ligand and receptor interactions might be either simply adhesive, graded along the abundance of receptors and affinity of interaction with target molecules, or involve a signalling component such that the protozoan receives information regarding the target cell which triggers a response, such as a change in flagellar activity. Alveolates possess a great range of possible carbohydrate-binding domains that appear to either be specific to classes within alveolates, or have broader distribution within prokaryotes and eukaryotes. Some domains are well-described, such as the conserved domains lectin, ricin and chitin binding (see Table 1); whereas many lectin proteins were identified based upon experimental affinity for carbohydrates as, for example, in Cryptosporidium (Bhat et al. Reference Bhat, Joe, PereiraPerrin and Ward2007; Bhalchandra et al. Reference Bhalchandra, Ludington, Coppens and Ward2013). Examples of carbohydrate-binding receptors include recognition of erythrocyte surface sialic acid by the Plasmodium merozoite protein EBA-175 during invasion (reviewed in Gaur and Chitnis, Reference Gaur and Chitnis2011); recognition of sialic acid via an unrelated saccharide-binding module, as well as a gal-lectin domain, within MIC1 participating in host cell invasion by Toxoplasma (TGME49_291890 in Fig. 2A; Friedrich et al. Reference Friedrich, Santos, Liu, Palma, Leon, Saoros, Kiso, Blackman, Matthews, Feizi and Soldati-Favre2010); and recognition of host carbohydrates mediated by an N-terminal domain within the microneme-secreted proteins MIC3 and MIC8 (CBL domain shown in TGME49_286740, Fig. 2A) during host cell invasion by Toxoplasma tachyzoites (Céréde et al. Reference Céréde, Dubremetz, Soête, Deslée, Vial, Bout and Lebrun2005). These carbohydrate-binding domains appear to be specific to 1 or more apicomplexan genera and are not found in proto-apicomplexans. Select alveolate proteins with predicted carbohydrate-binding activity are shown in Fig. 2A. It is probable that many alveolate carbohydrate-binding domains remain anonymous because they are not similar to known modules with such activity.
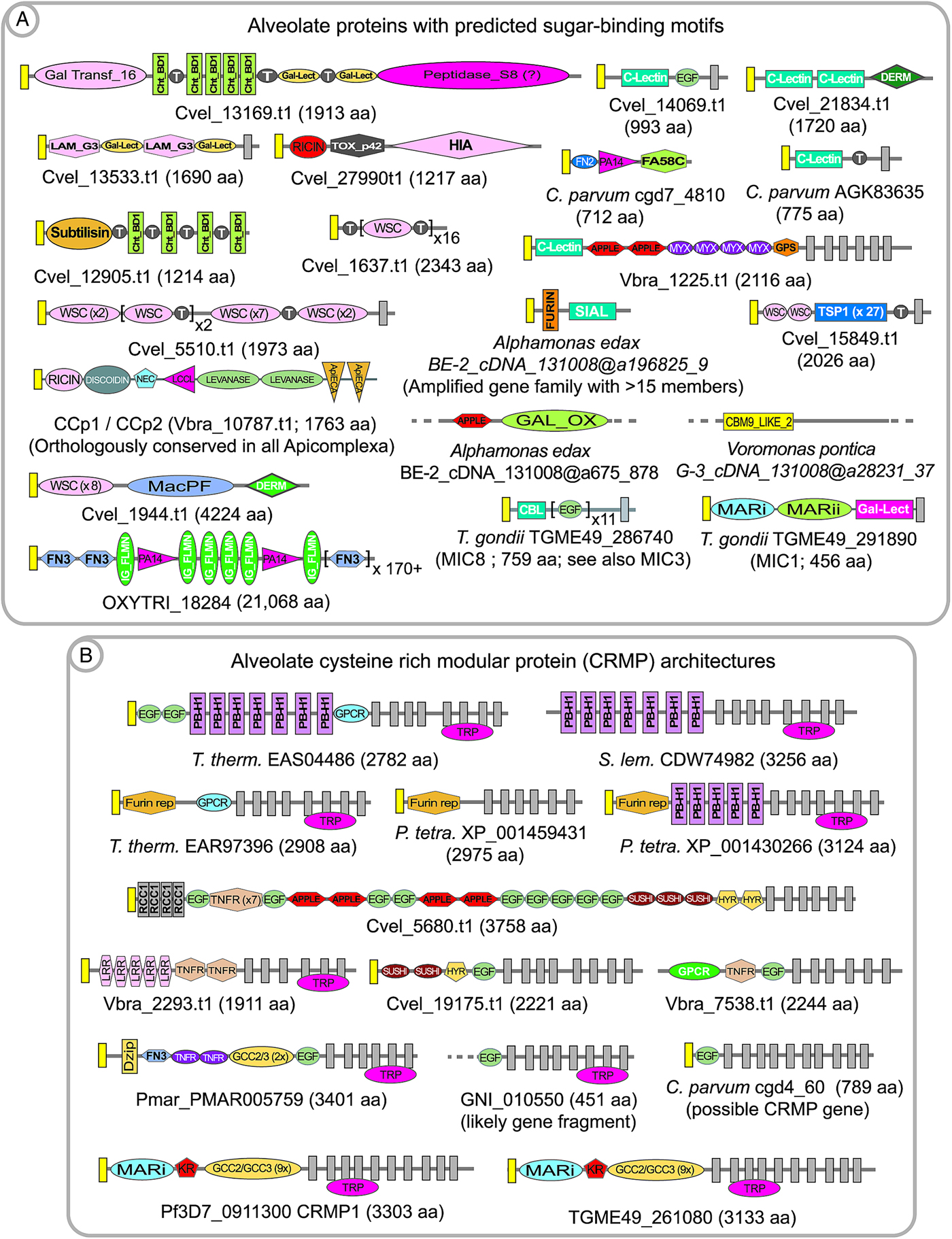
Fig. 2. Examples of predicted alveolate extracellular proteins having sugar-binding domains or mucin-like threonine repeat domains (A) and cysteine-rich modular protein (CRMP) architectures (B). Signal peptides are indicated by yellow boxes; and transmembrane domains are indicated by dark grey boxes. Colpodellids also possess CRMP proteins, but are not shown here due to the fragmentation of the transcriptome databases, and because they do not differ remarkably to chromerid examples. Domain abbreviations refer to either SMART (http://smart.embl-heidelberg.de/browse.shtml) or pfam (http://pfam.xfam.org) conserved domains. See also domain abbreviations and identifiers in Table 1.
Table 1. Phylogenetic distributions for select alveolate extracellular domains a

a White boxes indicate the absence and orange-shaded boxes indicate the presence of the domain (rows) in the genome or transcriptome sequence information for the relevant group (columns). Boxes shaded with grey and having a question mark indicate that the domain was not found, but the absence is qualified by the fact that the genome sequence information is incomplete for the relevant group.
bDomain accession identifiers. Domain information can be retrieved at the NCBI Conserved Domain website: http://www.ncbi.nlm.nih.gov/cdd.
cSpecies abbreviations: P. mar., Perkinsus marinus; C. vel., Chromera velia; V. brass., Vitrella brassicaformis; and C. parv., Cryptosporidium parvum.
dGeneral colpodellid grouping which includes Colpodella angusta, Colpodella_sp_BE-6, Alphamonas edax and Voromonas pontica. Domains were surveyed by local tblastn screening of transcriptome information (databases described in Janouškovec et al. Reference Janouškovec, Tikhonenkov, Burki, Howe, Kolísko, Mylnikov and Keeling2015) using chromerid and apicomplexan domain queries. The databases are incomplete and thus negative results are provisionally indicated by grey and a question mark, rather than white shading. Moreover, positive hits were not necessary for all organisms; for example, the HINT domain was only observed in V. pontica.
eAt the time of publication this accession identifier was valid, but the relevant entry could not be retrieved at the NCBI Conserved Domain website: http://www.ncbi.nlm.nih.gov/cdd.
fCysteine-rich domain found in Cryptosporidium oocyst wall proteins (COWP) and coccidians (Spano et al. Reference Spano, Puri, Ranucci, Putignani and Crisanti1997).
gDomain described in the Supplemental material for Templeton et al. (Reference Templeton, Iyer, Anantharaman, Enomoto, Abrahante, Subramanian, Hoffman, Abrahamsen and Aravind2004b ); specifically, ‘Domain typically with 6 cysteines, seen thus far mainly in animals with a few occurrences in plants. It is found in the sea anemone toxin metridin and fused to animal metal proteases, plant prolyl hydroxylases and is vastly expanded in the genome of C. elegans.’
Alveolate interactions with host cells might conversely involve purposeful display of carbohydrate residues on the parasite membrane surface, in order to engage host lectins. Such ‘mucin-like’ proteins are typically highly decorated with O-linked glycosylation, such as within large stretches of threonine and serine residues. One of the first apicomplexan mucin proteins to be identified was the Cryptosporidium sporozoite surface protein gp900 (e.g. AAC98153), which is proposed to be involved in host cell invasion (Barnes et al. Reference Barnes, Bonnin, Huang, Gousset, Wu, Gut, Doyle, Dubremetz, Ward and Petersen1998). Gp900 is composed of cysteine-rich domains and a lengthy array of threonine residues. Subsequent annotation of the C. parvum genome revealed a large repertoire of predicted mucin proteins (Abrahamsen et al. Reference Abrahamsen, Templeton, Enomoto, Abrahante, Zhu, Lancto, Deng, Liu, Widmer, Tzipori, Buck, Xu, Bankier, Dear, Konfortov, Spriggs, Iyer, Anantharaman, Aravind and Kapur2004), which are predominantly species-specific; in that for the most part they are not conserved in the repertoire of predicted mucins encoded within the genome of the Cryptosporidium parasite of gastric tissue, C. muris (Templeton, Reference Templeton, Ortega-Pierres, Cacciò, Fayer, Mank, Smith and Thompson2008). A mucin in Toxoplasma gondii, termed CST1 (TGME49_064660), is composed of a large repeat of SAG-related sequence (SRS) domains plus a threonine-rich array similar to Gp900, and is thought to be highly modified with N-acetyl-galactosamine (Tomita et al. Reference Tomita, Bzik, Ma, Fox, Markillie, Taylor, Kim and Weiss2013). CST1 was recently found to be crucial for the integrity of tissue cyst walls, with the threonine-rich region playing a critical role. Annotation of chromerids and colpodellids also reveals mucin proteins (for example Cvel_819.t1, Cvel_541.t1, and Colpodella_angusta_Spi-2_cDNA_ca@a28207_52), as well as a conserved O-linked glycosylation machinery which was first described in coccidians (Templeton et al. Reference Templeton, Iyer, Anantharaman, Enomoto, Abrahante, Subramanian, Hoffman, Abrahamsen and Aravind2004b ; Walker et al. Reference Walker, Slapetova, Slapeta, Miller and Smith2010). Our rough annotation of P. marinus indicates that it has perhaps an order of magnitude more genes which encode predicted mucins than Chromera, with potentially over 500 mucin genes within several families; based upon the features of predicted secretion, the presence of threonine repeats, and transmembrane or GPI-anchor domains. A Perkinsus mucin protein family has a conserved cysteine at the C-terminal residue, which possibly confers association with the surface membrane via fatty acylation of the cysteine residue (e.g. pmar_XP_002783417·1). Annotation of Chromera, Vitrella and colpodellids also revealed numerous proteins with predicted sugar-binding domains interspersed with threonine-rich repeats, suggesting that the proteins participate in polymerization to form intra- and inter-molecular matrices based upon sugar-binding motifs and sugar moieties (Fig. 2A). Parenthetically, regarding the stabilization of protein matrices, it has also been proposed that peroxidase-mediated cross-linking of di-tyrosine residues contribute to the integrity of coccidian oocyst walls (Mai et al. Reference Mai, Smith, Feng, Katrib, Slapeta, Slapetova, Wallach, Luxford, Davies, Zhang, Norton and Belli2011).
TWO COMPONENT SENSORY TRANSDUCTION HISTIDINE KINASE
Little is known regarding signal transduction across alveolate surface membranes in response to external environmental information, either in cell–cell or cell–nutrient interactions. The complexity of such signalling might distinguish apicomplexans and non-apicomplexans, if the former are considered to reside in relatively defined environments within hosts, and thus have a lesser requirement to respond to changing external environments during free-living life cycle stages. This hypothesis, however, must be reconciled with the apparent complexity of environmental recognition that arises during transformation between stages, changes in tissue localization within hosts, and transmission between hosts, such as during completion of the life cycle of Plasmodium.
One example of a possible alveolate signalling system, which is known only from annotation work and has not been pursued at the lab bench, is the observation that ciliates, Perkinsus and the chromerids possess large families of predicted two component sensory transduction histidine kinases (e.g. Cvel_8519.t1 in Chromera and Pmar_PMAR009211 in Perkinsus). The colpodellid transcriptome libraries also possess a broad range of 2 component sensory transducers, but these must be approached with caution due to possible bacterial contamination within the databases. In alveolates, these proteins possess multiple transmembrane domains, typically clustered at the N-terminus; a PAS domain (Pfam:PF00989) in some versions; a histidine kinase domain; and a C-terminal response receiver domain. The proteins appear to lack signal peptides, although the N-terminal transmembrane domain might function as a transfer sequence. In prokaryotes, the two component systems are integrated in the bacteria membrane and transduce a variety of environmental signals, but in alveolates their cellular localization and function has not been determined. In protozoans, the two component receptors are not exclusive to alveolates and are also found in stramenopiles, fungi and plants; and thus their origin in alveolates might have arisen through vertical inheritance. These receptors are absent in Apicomplexa; perhaps because their function, albeit unknown in alveolates, became vestigial following commitment to obligate parasitism.
CYSTEINE-RICH MODULAR PROTEIN (CRMP)
Annotation of predicted extracellular proteins within the whole genome sequence information for Plasmodium revealed an amplified gene family with 4 members, each encoded protein having a structural theme of large arrays of a cysteine-rich modules in the extracellular domain; multiple transmembrane spanning domains; and a large, low complexity predicted cytoplasmic domain (Thompson et al. Reference Thompson, Fernandez-Reyes, Sharling, Moore, Eling, Keyes, Newbold, Kafatos, Janse and Waters2007; Douradinha et al. Reference Douradinha, Ausustijn, Moore, Ramesar, Mota, Waters, Janse and Thompson2011). In addition to the cysteine-rich modules a single EGF-like domain, and in some versions an additional kringle domain were also present, leading to the name cysteine-rich modular protein (CRMP) for the family (e.g. PF3D7_0911300, PF3D7_1475400, PF3D7_0718300 and PF3D7_1208200 in P. falciparum). The recent description of a sialic-acid-binding module in some Toxoplasma microneme proteins (Friedrich et al. Reference Friedrich, Santos, Liu, Palma, Leon, Saoros, Kiso, Blackman, Matthews, Feizi and Soldati-Favre2010) allows identification of a similar domain in the N-terminal region of apicomplexan CRMPs. Gene knockout studies in Plasmodium demonstrated that the genes are essential for transmission to mosquitoes, and the protein products appear to function in the transmission stages (Thompson et al. Reference Thompson, Fernandez-Reyes, Sharling, Moore, Eling, Keyes, Newbold, Kafatos, Janse and Waters2007; Douradinha et al. Reference Douradinha, Ausustijn, Moore, Ramesar, Mota, Waters, Janse and Thompson2011). CRMPs are now known to be present in all apicomplexans, with the exception of Cryptosporidium, as well as Perkinsus and are amplified in large families in the chromerids, colpodellids and ciliates (Fig. 2B). They also have a broader distribution in protozoa, such as stramenopiles, suggesting their presence in the last common ancestor of alveolates. The multi-transmembrane region is conserved across the phylogenetic distribution and often shows similarity to an ion channel termed the transient receptor potential (TRP) domain (reviewed in Venkatachalam and Montell, Reference Venkatachalam and Montell2007). Thus a reasonable hypothesis is that the CRMP proteins participate in an environmental sensing role, with extracellular recognition of ligands and signalling across the membrane.
In chromerids, CRMP proteins are highly amplified, with approximately 40 members in Chromera. The fragmented nature of the colpodellid transcriptome libraries precludes an estimation of the extent of the gene amplifications, but they appear to have similar domain structures to examples in the chromerids. The ciliates Tetrahymena, Oxytricha and Stylonychia also have a highly amplified representation of CRMP proteins, with up to 100 genes within each genome. Thus, a great reduction in the number and variety of CRMP proteins accompanied the transition to the apicomplexan clade, with a complete loss of the genes in Cryptosporidium. This is perhaps in accordance with a role of CRMP proteins in recognition and response to the extracellular environment; one hypothesis being that the obligate parasitic apicomplexans might encounter a relatively defined environment, and thus do not require a broad repertoire of CRMP proteins. Perkinsus also appears to have a reduced number of CRMP proteins, and thus a correlation of CRMP proteins with life strategies would need to take into account the parasitic and free-living components of the Perkinsus life cycle; but also indicates a correlation with parasitism and loss of crmp genes.
CAST MULTI-DOMAIN PROTEIN
Numerous alveolates possess members of an amplified gene family, termed CAST multi-domain protein, which has not been functionally characterized. These giant proteins, ranging from several thousand amino acids in length, have architectures consisting of a large repeated array of cysteine-rich modules; a single transmembrane domain having conserved features; and a large (>150 kDa) presumed cytoplasmic domain having a low complexity, predicted coiled-coil character. The protein probably originated prior to the divergence of the alveolate lineage, since it is also found in stramenopiles and choanoflagellates. In the ciliate Oxytricha, the gene is highly amplified, with perhaps over 100 members (e.g. OXYTRI_15408); whereas in Toxoplasma there appear to be less than 5 genes encoding predicted CAST multi-domain proteins (e.g. TGME49_207480 and TGME49_253930), which are typically annotated as ‘GCC2 and GCC3 domain-containing proteins.’ Across the alveolates, apparent gene losses have shaped the phylogenetic distribution of the gene, and within the alveolates representatives of this protein are present in the ciliate Oxytricha, but not in Tetrahymena and Paramecium; in Chromera (e.g. Cvel_3066.t1), Vitrella and colpodellids, but absent in Perkinsus; in the coccidians, Eimeria and Toxoplasma; and absent in other apicomplexans such as Cryptosporidium, Theileria, Babesia and Plasmodium. The conserved sequence surrounding the transmembrane region suggests a conservation of a juxta-membrane function, such as interaction with the membrane or signalling. The cytoplasmic domain of the CAST multi-domain protein, which includes the namesake (and perhaps erroneously ascribed) CAST domain, appears to be conserved and is large (over 1500 aa), low complexity and with possibly with a coiled-coil structure. Toxoplasma is the obvious experimental organism in which to determine the cellular localization and function of the CAST multi-domain proteins.
OOCYST WALL PROTEIN
Cryptosporidium oocysts can be obtained in abundance and high purity following the experimental infection of a calf. Thus, Cryptosporidium is an excellent system in which one can study coccidian cyst structure (Spano et al. Reference Spano, Puri, Ranucci, Putignani and Crisanti1997; Chattrejee et al. Reference Chattrejee, Banerjee, Steffen, O'Conner, Ward, Robbins and Samuelson2010; Samuelson et al. Reference Samuelson, Bushkin, Chatterjee and Robbins2013). An oocyst wall protein, termed OWP or Cryptosporidium oocyst wall protein (COWP), was purified from oocyst wall extracts and its protein sequence determined (Spano et al. Reference Spano, Puri, Ranucci, Putignani and Crisanti1997). The COWP protein is composed of repeats of variations of a highly cysteine-rich module. With the advent of whole genome sequence information it is now known that COWP genes are amplified in Cryptosporidium, which has 9 genes, and are also amplified in all cyst-forming coccidians and Gregarina (Fig. 3A; Templeton et al. Reference Templeton, Lancto, Vigdorovich, Liu, London, Hadsall and Abrahamsen2004a ; Templeton et al. Reference Templeton, Enomoto, Chen, Huang, Lancto, Abrahamsen and Zhu2010). OWP modules are also found in genes amplified in the chromerids (e.g. Vbra_11165.t1 in Vitrella and Cvel_20950.t1 in Chromera; Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015) and colpodellids, and thus, this component of the structure of the oocyst predates the specialization to Apicomplexa. OWP genes are not found in Perkinsus, dinoflagellates and ciliates, and thus their origin possibly occurred in the last common ancestor of the chromerids and apicomplexans. It has not been addressed if specific COWP genes are orthologously shared in the chromerids and colpodellids; nor if it is known if genes are vertically inherited as orthologues in apicomplexans, thus indicating possible conserved functions within the oocyst wall structure. The genes are differentially amplified in Chromera versus Vitrella, and this might indicate that differing architectures underpin differing oocyst wall characters in the 2 closely related protozoans. Vitrella possesses as many as 30 OWP genes, whereas Cryptosporidium possesses 9 genes, emphasizing possible structural differences related to the number of encoded genes. Apicomplexans which do not have an externally shed oocyst stage have lost OWP genes; such as Plasmodium, Babesia and Theileria. Perkinsus lacks OWP genes and also does not possess a durable cyst stage; with only one report describing an apparently abundant ‘cell wall’ protein which is probably unrelated to cyst walls (Montes et al. Reference Montes, Durfort, Llado and Garcia-Valero2002). The observation that OWP proteins are present in proto-apicomplexans provides markers with which to describe the great diversity in structures of inner and outer cell walls in the alveolates.
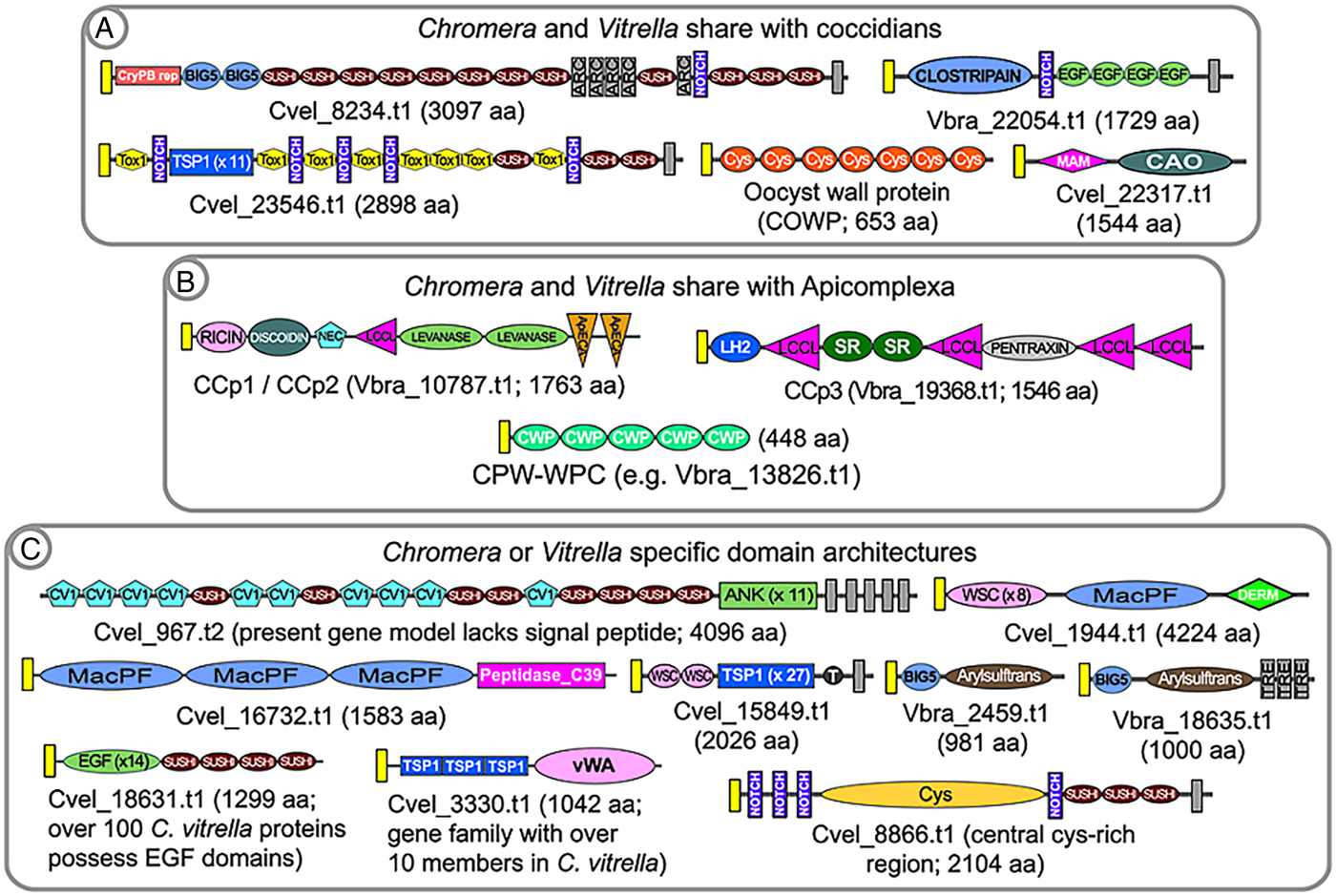
Fig. 3. Examples of predicted alveolate extracellular proteins that unite Chromera and Vitrella to coccidians (A) or Apicomplexa (B), to the exclusion of ciliates and dinoflagellates. Because of the fragmented nature of the colpodellid databases it is not possible to include them in these analyses; rather, they are discussed in the text. Examples of Chromera or Vitrella-specific domain architectures are shown in (C). Signal peptides are indicated by yellow boxes; and transmembrane domains are indicated by dark grey boxes. Domain abbreviations refer to either SMART (http://smart.embl-heidelberg.de/browse.shtml) or pfam (http://pfam.xfam.org). See also domain abbreviations and identifiers in Table 1.
CHROMERIDS AND COLPODELLIDS AS COCCIDIANS
The conservation of the OWP in chromerids, colpodellids and coccidians, but their absence in Perkinsus and ciliates, is congruent with the known taxonomic affinity of chromerids and colpodellids with the Apicomplexa. Annotation of the predicted proteome of the chromerids revealed numerous additional predicted extracellular proteins having complex, multi-domain architectures which are shared with coccidians (Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015). Examples include the large, multi-domain protein TRAP-C2, first described in Cryptosporidium (perhaps erroneously named and not a TRAP family protein; Spano et al. Reference Spano, Putignani, Naitza, Puri, Wright and Crisanti1998); a protein with a fusion of a MAM domain and a copper amine oxidase; and a transmembrane protein containing clostripain, notch and EGF domains (Fig. 3A). A first hypothesis might be that these proteins are involved in formation of the coccidian external oocyst, since they are not present in other apicomplexans, such as Plasmodium and Babesia, which lack external cyst stages; nor are they conserved in ciliates or Perkinsus. Thus, as suggested from phylogenetic trees, the last common ancestor of the apicomplexan lineage was coccidian-like; in that it possessed an environmentally-durable cyst stage. Two proteins, one with a HINT domain (e.g. C. parvum, cgd7_5290; Gregarina, GNI_039770; and Chromera, Cvel_10247.t1) and another encoding Fringe + Galactose transferase (e.g. C. parvum, cgd6_1450; and Chromera, Cvel_3306.t1), group the chromerids with Cryptosporidium or Gregarina to the exclusion of the coccidians and other Apicomplexa, thus providing support for placing Gregarina and Cryptosporidium at the base of the apicomplexan clade.
For colpodellids, it remains difficult to identify predicted orthologues of multi-domain extracellular proteins because the transcriptome databases are fragmented. For example, we have identified many copper amine oxidases in the colpodellid databases, but no fusions with a MAM domain, as described above. Another example is the presence of possible fragments, but no full-length TRAP-C2 orthologues. For this reason, it is of value to annotate colpodellid transcriptomes for the presence or absence of component extracellular domains, rather than survey for orthologues of large, complex multi-domain proteins. Here again the chromerids share extracellular domains with coccidians, to the exclusion of colpodellids; for example, the MAM domain as described above; a clostripain domain, found fused to Notch and EGF repeats in 1 coccidian and chromerid protein; and the TOX1 domain (Table 1). However, it is important to obtain complete genome sequence information for one or more colpodellids, because the transcriptome information might not have sufficiently high coverage, particularly across life cycle stages, for discussions of negative data.
CHROMERIDS AND COLPODELLIDS AS APICOMPLEXANS
Annotation of the P. falciparum genome revealed a family of proteins, termed CCP or LAP, having a rich multi-domain architecture of predicted sugar and lipid-binding domains (Pradel et al. Reference Pradel, Hayton, Aravind, Iyer, Abrahamsen, Bonawitz, Mejia and Templeton2004; Raine et al. Reference Raine, Ecker, Mendoza, Tewari, Stanway and Sinden2007; Carter et al. Reference Carter, Shimuzu, Arai and Dessens2008). Studies in P. falciparum and the rodent malaria parasite, Plasmodium berghei, indicate that the proteins function in sexual stage parasites, and gene disruption studies indicate a probable manifestation of phenotype in the mosquito midgut ookinete stage. Recent whole genome information indicates that the CCP/LAP genes are conserved as homologues not only across Apicomplexa, including Cryptosporidium and Gregarina, but also in the chromerids, Chromera and Vitrella (see Fig. 3 – figure supplement 4 in Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015). The colpodellids also possess the component domains of CCP/LAP proteins, although the fragmentation of the transcriptome sequence information makes determination of possible orthologous conservation of the multi-domain architectures. All members of the CCP/LAP family, as well as their component domains, are absent in ciliates and Perkinsus. Thus, any hypotheses of their function in Apicomplexa must also consider their function to be ancient, with orthologues present in the chromerids. The CCP/LAP proteins are predicted to be targeted to the crystalloid of Plasmodium ookinetes (Carter et al. Reference Carter, Shimuzu, Arai and Dessens2008), in addition to extracellular secretion, and thus might serve as markers to determine if a similar organelle is present in all apicomplexans and chromerids. The cysteine-rich CPW_WPC domain family additionally present in all apicomplexans, with the exception of Cryptosporidium; is amplified in the chromerids and colpodellids; and is absent in Perkinsus and the ciliates. This protein family thus represents another marker with which to investigate conserved structures uniting proto-apicomplexans and apicomplexans.
The phylogenetic distribution of the component domains of predicted extracellular multi-domain proteins also group the chromerids with either ciliates, ciliates plus Perkinsus, coccidians, apicomplexans or all alveolates (Table 1). For example, component domains of the multi-domain CCP/LAP proteins (namely, ricin, NEC, SR, LCCL, as well as other domains) also have a phylogenetic distribution uniting the chromerids with Apicomplexa, to the exclusion of Perkinsus and the ciliates. Many of the extracellular domains common to chromerids and Apicomplexa are also found in metazoans, and thereby may have arisen through lateral transfer (Templeton et al. Reference Templeton, Iyer, Anantharaman, Enomoto, Abrahante, Subramanian, Hoffman, Abrahamsen and Aravind2004b ; Aravind et al. Reference Aravind, Anantharaman, Zhang, de Souza and Iyer2012). Table 2 illustrates the variety of domains and multi-domain architecture expansions, using chromerids as examples.
Table 2. Examples of Vitrella and Chromera multi-gene families encoding predicted secreted proteins and component domains

a Number of genes in Vitrella with a specific domain, or the number of paralogues within the relevant gene family. Some proteins have repeats of a specific domain, such as arrays of TSP1, Sushi or COWP domains.
Not shown in Tables 1 and 2 are the numerous alveolate extracellular domains and proteins which appear to have been ‘invented’ de novo, in that they are genera- or species-specific. Some of these are discussed elsewhere in this review, such as presumptive saccharide-binding domains, and examples of the numerous highly amplified, anonymous protein families in the ciliates and dinoflagellates. Within P. falciparum examples of lineage-specific domains and proteins include the Duffy binding-like domain within PfEMP1 and EBA-175-like proteins which confer cytoadhesion of infected erythrocytes and recognition of erythrocyte during invasion, respectively; and the SURFIN, RIF and STEVOR proteins, which have unknown functions (Dzikowski et al. Reference Dzikowski, Templeton and Deitsch2006; Frech and Chen, Reference Frech and Chen2013). Parasite-encoded erythrocyte surface proteins also show species-specificity; for example, the SICAvar proteins found in Plasmodium knowlesi and other primate malaria parasites (al-Khedery et al. Reference al-Khedery, Barnwell and Galinski1999; Frech and Chen, Reference Frech and Chen2013; Lapp et al. Reference Lapp, Korir-Morrison, Jiang, Bai, Corredor and Galinski2013). Other genera-specific examples of domains and proteins are the highly amplified families of secreted FAINT domain proteins in Theileria (Pain et al. Reference Pain, Renauld, Berriman, Murphy, Yeats, Weir, Kerhornou, Aslett, Bishop, Bouchier, Cochet, Coulson, Cronin, de Villiers, Fraser, Fosker, Gardner, Goble, Griffiths-Jones, Harris, Katzer, Larke, Lord, Maser, McKellar, Mooney, Morton, Nene, O'Neil, Price, Quail, Rabbinowitsch, Rawlings, Rutter, Saunders, Seeger, Shah, Squares, Squares, Tivey, Walker, Woodward, Dobbelaere, Langsley, Rajandream, McKeever, Shiels, Tait, Barrell and Hall2005) and the VESA erythrocyte surface antigens in Babesia (O'Conner et al. Reference O'Conner, Lane, Stroup and Allred1997; Jackson et al. Reference Jackson, Otto, Darby, Ramaprasad, Xia, Echaide, Farber, Gahlot, Gamble, Gupta, Gupta, Jackson, Malandrin, Malas, Moussa, Nair, Reid, Sanders, Sharma, Tracey, Quail, Weir, Wastling, Hall, Willadsen, Lingelbach, Shiels, Tait, Berriman, Allred and Pain2014). In coccidians, the SAG and SRS proteins are perhaps the best examples of ‘inventions’ conferring host interactions, in this instance likely having an origin via lateral gene transfer, as described in a previous section. Such highly evolved lineage-specific proteins may have conferred new host interactions which allowed exploitation by the parasite, followed by selection by functional and host immune response pressures which drove their diversification and amplification (for reviews see, e.g., Templeton, Reference Templeton2009; Mackinnon and Marsh, Reference Mackinnon and Marsh2010; Smith et al. Reference Smith, Rowe, Higgins and Lavstsen2013; Jackson et al. Reference Jackson, Otto, Darby, Ramaprasad, Xia, Echaide, Farber, Gahlot, Gamble, Gupta, Gupta, Jackson, Malandrin, Malas, Moussa, Nair, Reid, Sanders, Sharma, Tracey, Quail, Weir, Wastling, Hall, Willadsen, Lingelbach, Shiels, Tait, Berriman, Allred and Pain2014; Smith, Reference Smith2014).
ON THE ORIGIN OF GLIDING MOTILITY IN APICOMPLEXANS
Arguably the singular revolution in the transition to obligate parasitism in the Apicomplexa was the development of gliding motility as a means to facilitate tissue traversal and cell invasion. Alveolates use flagella for motility, typically as flagella pairs in the case of the dinoflagellates and chromerids; in rows of cilia, such as in the namesake ciliates; or the combination of cilia and single apical flagella, such as described in the elegant Ileonema simplex. Gliding motility, however, is unique to apicomplexans, although flagellar motility has been retained in the microgamete stages of Plasmodium. Apicomplexan gliding motility has been well described elsewhere (e.g. Daher and Soldati-Favre, Reference Daher and Soldati-Favre2009; Frénal et al. Reference Frénal, Polonais, Marq, Stratmann, Limenitakis and Soldati-Favre2010; Jacot et al. Reference Jacot, Frénal, Marq, Sharma and Soldati-Favre2014); and discussion of the intracellular components of the gliding motility molecular machinery, termed the glideosome, in proto-apicomplexans are described in Woo et al. (Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015). Here we will describe the contribution of new genome sequence information to understanding the possible origin of the apicomplexan surface receptor involved in gliding motility, called the TRAP/MIC2 superfamily of transmembrane proteins (reviewed in Morahan et al. Reference Morahan, Wang and Coppel2009).
TRAP/MIC2 family proteins link extracellular adhesion to interaction with the cytoplasmic actin and myosin motility apparatus. All TRAP/MIC2 proteins described to date possess an extracellular region containing one or more TSP1 domains and, in many instances, one or multiple vWA domains. Additional hallmarks of TRAP/MIC2 proteins are a single transmembrane domain, in some instances with a juxta-membrane rhomboid protease cleavage site; a short, charged cytoplasmic domain; and C-terminal region aromatic residues which are thought to interact with cytoplasmic components of the motility apparatus. Figure 4A shows the variety of domain architectures from predicted TRAP/MIC2 members across the Apicomplexa. The chromerids, which appear to lack gliding motility, possess numerous predicted extracellular proteins harbouring TSP1 and vWA domains; thus the presence of these domains in the alveolates does not correlate with gliding motility. Indeed, Vitrella has an expansion of over 30 proteins harbouring TSP1 domains. Vitrella, Chromera and the colpodellid A. edax all possess proteins with 3 TSP1 domains followed by a C-terminal vWA domain; and the proteins appear to have an orthologous relationship based upon the presence of additional conservation throughout the sequence to the N-terminal side of the TSP1 domains (Fig. 4A). Thus, this TSP1 plus vWA domain architecture probably has a conserved function in the colpodellids and chromerids. However, none of these proteins appear to possess the additional hallmark TRAP/MIC2 features; namely, a transmembrane domain followed by a short, charged cytoplasmic domain having aromatic residues (qualifying here that gene models may not have been not precisely determined for the chromerids and colpodellids).

Fig. 4. Domain architectures of predicted apicomplexan TRAP proteins and representative TSP1 and vWA proteins from chromerids, colpodellids and Gregarina (A). Proteins are not drawn to scale; lengths in amino acids (aa) indicated. The gene for Alphamonas edax BE-2 _cDNA_131008@a34668_32 appears to be incomplete at the 3′ end, indicated by a dashed line. Amino acid sequences of the cytoplasmic domains of TRAP/MIC2 proteins and candidates; the sequences are not aligned based upon amino acid similarities, but rather to show conserved features within the short, acidic cytoplasmic domain and conserved aromatic residues adjacent the C-terminus (B). Predicted transmembrane regions are highlighted in blue and aromatic residues highlighted in yellow. Gene IDs are as follows: PfCTRP (PF3D7_1133400), PfTRAP (PF3D7_1133400), TgMIC2 (TGME49_001780), CpTSP1 (cgd1_3500), CpTSP7 (cgd5_4470), GnvWA (GNI_102870 and GNI_030200), GnTSP (GNI_006920 and GNI_113530).
Broad coverage genome sequence information has recently become available for the apicomplexan, G. niphandrodes, in which gliding motility is well described. Gregarines possess exquisite drapery-like longitudinal surface structures termed epicytic folds, which are proposed to be involved in gliding motility (reviewed in Valigurová et al. Reference Valigurová, Vaškovičová, Musilová and Schrevel2013). We were unable to identify clear homologues of TRAP/MIC2 proteins in the G. niphandrodes; however, the parasite does possess numerous genes encoding proteins having single vWA domains, including examples with signal peptides, C-terminal transmembrane domains and C-terminal aromatic residues (Fig. 4A and B). The gene predictions for G. niphandrodes appear to be preliminary and require validation, but the number may exceed 20 such TRAP-like proteins. The cousin of gregarines, Cryptosporidium, has multiple predicted TRAP/MIC2 proteins, although this protozoan lacks extracellular examples of vWA domains; rather, the Cryptosporidium predicted TRAP/MIC2 proteins are composed of TSP1 and apple domains (Deng et al. Reference Deng, Templeton, London, Bauer, Schroeder and Abrahamsen2002). One Cryptosporidium TSP1 domain protein, termed TRAP-C2, has a large array of TSP1 domains, plus Notch, TOX1 and CCP/Sushi domains, and a C-terminal transmembrane domain. However, this protein does not have TRAP/MIC2 features within the predicted cytoplasmic domain; namely, a charged character and C-terminal aromatic residues. TRAP-C2 is now known to be conserved as predicted orthologues in coccidians, gregarines, as well as chromerids (Fig. 3A; in Chromera, Cvel_23546.t1). The G. niphandrodes version differs in that the predicted cytoplasmic domain is charged and possesses C-terminal aromatic residues, and thus might be investigated as a candidate TRAP protein. The Cryptosporidium protein GP900, discussed above, has been implicated in cell invasion and is composed of extracellular arrays of a genera-specific, cysteine-rich domain; a single transmembrane domain; and a short, charged cytoplasmic domain with aromatic residues reminiscent of TRAP/MIC2 proteins. Tissue culture is unavailable for G. niphandrodes and C. parvum and thus limits genetic manipulation; however, a newly developed mouse model and gene manipulation method for C. parvum (Vinayak et al. Reference Vinayak, Pawlowic, Sateriale, Brooks, Studstill, Bar-Peled, Cipriano and Streipen2015) shows great promise and might be used to characterize the function of potential TRAP/MIC2 proteins. Alternatively, the ability of candidate proteins to complement TRAP/MIC2 proteins might be tested in another system, such as Toxoplasma. If GP900 functions as a TRAP/MIC2 protein, despite its lack of TSP1 or vWA domains, then this would indicate that the prototypic features of a gliding motility receptor might be the TRAP/MIC2-like cytoplasmic domain. The TRAP/MIC2 architectural paradigm works well in identifying apicomplexan receptor candidates for mediating gliding motility, but possible proto-apicomplexan precursors to such proteins remain obscure since clear orthologous relationships are not apparent.
What can be said about the chromerids and colpodellids, as representative proto-apicomplexans, with respect to the innovation of gliding motility? One-to-one orthologous relationships of the intracellular components of glideosome proteins have not been conclusively identified (Woo et al. Reference Woo, Ansari, Otto, Klinger, Kolisko, Michálek, Saxena, Shanmugam, Tayyrov, Veluchamy, Ali, Bernal, del Campo, Cihlář, Flegontov, Gornik, Hajdušková, Horák, Janouškovec, Katris, Mast, Miranda-Saavedra, Mourier, Naeem, Nair, Panigrahi, Rawlings, Padron-Regalado, Ramaprasad, Samad, Tomčala, Wilkes, Neafsey, Doerig, Bowler, Keeling, Roos, Dacks, Templeton, Waller, Lukeš, Oborník and Pain2015), but related protein expansions have been observed for GAP40, GAP50, GAPM and ISP proteins. These sequence similarities did not extend to the ciliates, which supports the phylogenetic relationship of chromerids and apicomplexans. Greater understanding of the origin of gliding motility may come from refining proteomic and molecular studies to characterize candidate proteins, as well as obtaining ultrastructural, proteomic and whole genome sequence and transcriptome information for more proto-apicomplexan organisms. Describing the evolution of TRAP/MIC2 proteins awaits a better understanding of the function of gregarine and Cryptosporidium predicted receptor proteins.
Concluding remarks
Recently derived whole genome sequence information for the chromerids, C. velia and V. brassicaformis, and high coverage transcriptome information for colpodellids, supports their phylogenetic relationship with the Apicomplexa, and allows annotation with the goal of describing the molecular hallmarks of transition of a free-living alveolate to obligate parasitism in the Apicomplexa. The annotations described herein support the hypothesis that chromerids are more closely related to Apicomplexa than are the alveolates Perkinsus, dinoflagellates and ciliates, and thus far serve as the closest and best-described ‘outgroup’ in which we can study the transition to parasitism in the Apicomplexa. However, the chromerids also possess highly amplified families of predicted external sensory proteins uniting them with the dinoflagellates and ciliates. The great reduction or complete loss of orthologues for these families within Apicomplexa suggests, as one hypothesis, that obligate parasitism reduces the requirement for response to interacting with and interpreting the unpredictable and highly variable external environment. Conservation of numerous predicted extracellular proteins, such as the OWP domain-containing oocyst wall proteins, as well as complex multi-domain proteins, between the chromerids and coccidians suggest that structural aspects of the cyst stage are conserved; that is, the chromerids can be viewed as ‘model coccidians’ rather than grouping with dinoflagellates or ciliates. Other conserved extracellular proteins, such as the LCCL and CPW_WPC domain containing proteins and numerous extracellular domains also group the chromerids with all apicomplexans. The chromerids have provided few clues towards understanding the development of gliding motility in the apicomplexans, although some glideosome proteins appear to have origins prior to the transition to Apicomplexa. Further functional and systems biology studies, such as in the gregarines and proto-apicomplexans, are required to unravel the steps which occurred in the evolution of gliding motility in the apicomplexans.
ACKNOWLEDGEMENTS
The authors would like to thank Bruce Taylor and Richard Culleton for their critical readings of the paper.
FINANCIAL SUPPORT
T.J. Templeton would like to acknowledge the generous support of a Visiting Professorship at the Institute of Tropical Medicine (NEKKEN), Nagasaki University, Japan. Research in A. Pain's research group is supported by KAUST-faculty baseline funding and CRG grants from OCRF, KAUST and Global Station for Zoonosis Control, Global Institution for Collaborative Research and Education (GI-CoRE), Hokkaido University, Japan.











