Introduction
Barley (Hordeum vulgare ssp. vulgare L.) is the crop with the longest documented history of domestication, dating back to around 10,000 years (Badr et al., Reference Badr, M, Sch, Rabey, Effgen, Ibrahim, Pozzi, Rohde and Salamini2000). Barley has been an important crop, ranking fourth among all cultivated crops worldwide, with a total surface of nearly 48 million hectares (Statista, 2022). Since it can be cultivated under quite variable geographic and climatic conditions and, thus, is able to survive and tolerate diverse stress conditions. Thus, barley is considered a promising candidate for a resilient crop (Newton et al., Reference Newton, Flavell, George, Leat, Mullholland, Ramsay, Revoredo-Giha, Russell, Steffenson, Swanston, Thomas, Waugh, White and Bingham2011) with great potential to safeguard food security under the challenge of global climate change (Leisner, Reference Leisner2020).
The importance of genetic diversity in crop plants and its conservation as future breeding resource has become a matter of global concern (Convention on Biological Diversity 2006, 2007). Barley is no exception, since the breeding of modern barley varieties went along with a progressive loss of genetic diversity (Bernardo, Reference Bernardo2014). At the same time, traditional landraces, endowed with superior adaptability and stress resilience (but with the drawback of lower yields, compared to the modern elite cultivars), have become rare and often even got extinct (Ceccarelli, Reference Ceccarelli, Maxted, Ehsan Dulloo, Ford-Lloyd, Frese, Iriondo and Pinheiro de Carvalho2012). Hence, the investigation on barley biodiversity has become essential to identify and subsequently to maintain these genetic resources with their promising potential. Prominently, the integration of such landraces into future improvement strategies will be crucial to meet the production and agricultural needs of an agriculture that must cope with the challenges of global climate change. Introgression of resilience factors into high-yielding elite cultivars represents a new aim of current agricultural breeding and is facilitated by the use of molecular markers that allow for accelerated breeding (Al-Khayri et al., Reference Al-Khayri, Jain and Johnson2019).
The first step on the path towards introgression is the genetic characterisation of potential breeding sources, such as traditional varieties, landraces or even crop wild relatives (CWR). In this context, DNA-based molecular markers have been an efficient tool in the selection of desirable varieties and traits, they are widely used to distinguish or authenticate genotypes, but can also be used to infer the phylogenetic relations between them. Also, for barley, such markers have been used extensively. Small sequence repeat (SSR) or microsatellites have been in the focus (for review see Struss and Plieske, Reference Struss and Plieske1998), because they were developed in concert with the barley genome project (Liu et al., Reference Liu, Biyashev and Maroof1996), and thus also yield information on the relationship of specific chromosomes based on polymorphic differences in size. While both SSR and random amplifying polymorphic DNA (RAPD) markers are exclusively relying on marker length, also sequence-dependent approaches such as Sequence Tagged Sites (STS) are progressively gaining momentum. Each of these strategies has its benefits and drawbacks, as neatly worked out in a comparative study on 38 barley varieties from North America, Western and Eastern Europe (Meszaros et al., Reference Meszaros, Karsai, Kuti, Banyai, Lang and Bedo2007).
Since the location of SSR markers is known, they can be chosen in a manner that they represent chromosomes symmetrically, rendering them suitable to infer phylogenetic relationships. Instead, the position of RAPD markers is usually not known, and they are merely selected to differentiate the genotypes of interest by their polymorphism. Therefore, they are often considered as outdated, because they are less representative for phylogenetic relationships as compared to SSR markers. However, it is exactly this bias that can add insight – for instance, when closely related genotypes need to be compared. Here, RAPDs, focusing on the difference, allow for ‘zooming in’ and, thus, to differentiate otherwise very similar genotypes. This can be helpful during the investigation of geographic relationships as important aspect for the study of landraces. Therefore, RAPD markers have remained a valuable tool to analyse plant diversity in general (Bοusba et al., Reference Bοusba, Gueraiche, Kanouni, Bounar, Djekoune, Khammar and Ykhlef2020; Singh et al., Reference Singh, Kudesia and Bhadauria2020; Somana et al., Reference Somana, Saha, Rashid and Haque2021), as well as barley diversity in particular (Fadel et al., Reference Fadel, Abdulhamed and Yousif2022). Thus, the use of both, RAPD and SSR markers has to be considered as complementary strategies to address both, phylogenetic and geographic relationships within a crop (Mir et al., Reference Mir, Karmakar, Chattopadhayay, Chaudhury, Gosh and Roy2008).
In Algeria, barley used to be, traditionally, the leading cereal and still ranks third area wise with respect to distribution. The production and importance of barley usage have changed through time (Rahal-Bouziane, Reference Rahal-Bouziane2015), and were fundamentally influenced by the French colonisation (Laumont, Reference Laumont1937; Tounsi, Reference Tounsi1986). Although its importance as human food has declined somewhat, the use of barley as animal fodder has remained important. Barley malting, which predominates in Europe, is only of marginal importance in Algeria. Barley in Algeria is dominated by mixed cultivation of local varieties with genotypes introduced from the Near East. The varieties with the largest economic impact are Saïda183 and Tichedrett that derive from landraces adapted to semi-arid high-plateaus area (Ullrich, Reference Ullrich2011). They represent about 90 and 10% of the cultivated area, respectively, and are the varieties preferred by the farmers in Algeria (Reguieg et al., Reference Reguieg, Labdi, Benbelkacem, Hamou, Maatougui, Grando and Ceccarelli2013). In addition, several varieties were released in the frame of breeding programmes performed mainly in cooperation with the International Center for Agricultural Research in the Dry Areas (ICARDA), or bilateral collaboration programmes such as with the Institut National de Recherche Agronomique de Tunis (INRAT) and Institut National de Recherche Agronomique Maroc (INRAM). However, these introduced varieties did not perform successfully, showing high susceptibility to the climate conditions prevailing in Algeria (Benmahammed, Reference Benmahammed1996). Therefore, Saïda183 and Tichedrett are still the varieties most popular among the farmers in Algeria because of their forage type habit. In addition to their superior resilience, they also perform well with respect to nutritive values equalling modern European cultivars (Arbouche et al., Reference Arbouche, Arbouche, Arbouche and Arbouche2008).
Despite the intensive use of molecular genetics to understand barley biodiversity and to support barley breeding, the Western Mediterranean gene pool has been systematically neglected. This is not only valid for the barley varieties from Spain (Yahiaoui et al., Reference Yahiaoui, Igartua, Moralejo, Ramsay, Molina-Cano, Ciudad, Lasa, Gracia and Casas2008), but also holds true for the Algerian barley varieties. While several studies have screened the variability for Algerian barley accessions with respect to pheno-morphological and agro-economical selection and traits (Rahal-Bouziane et al., Reference Rahal-Bouziane, Merdas, Merzoug and Abdelguerfi2015, Reference Rahal-Bouziane, Yahiaoui, Merdas, Berkani, Nait-Merzoug and Abdelguerfi2020; Taibi et al., Reference Taibi, Bellatreche A and Gaouar2019; Bendada, Reference Bendada2021), the knowledge on the phylogenetic relationships among these varieties has remained limited, although there exist a few studies, based on SSR markers, which had included also Algerian accessions, either in the context with other North African accessions (Ben Naceur et al., Reference Ben Naceur, Chaabane, El-Faleh, Abdelly, Ramla, Nada, Sakr and Ben Naceur2012), or as part of a global screen for drought tolerance (Varshney et al., Reference Varshney, Baum, Guo, Grando, Ceccarelli and Graner2008, Reference Varshney, Salem, Baum, Roder, Graner and Börner2010). Thus, the scope of these studies was extending beyond Algerian germplasm. The same holds true for a study employing RAPD markers to characterise genetic diversity in North African accession of barley (Allel et al., Reference Allel, Ben-Amar, Lamine and Abdelly2017).
Barley cultivation in Algeria is still relying to a large degree on local landraces that had been improved by selection, and, during recent years, have been complemented by varieties introduced from Europe through the ICARDA programme. To what extent the identity of traditional varieties can be safeguarded, remains an open question. The current study tried to get insight into the autochthony of the traditional landraces Saïda183 and Tichedrett in relation to introduced varieties and their derivatives (since these introduced varieties are often cultivated in the same manner as traditional landraces, such that they are likely prone to evolutionary change). Methodologically, we chose a complementary approach, where phylogenetic relationships were addressed using SSR markers that symmetrically represented the different chromosomes, while the search for informative RAPD markers allowed insights into geographic relationships beyond mere genetics. In addition, a principal component analysis (PCA) of agrophenotypic traits linked the traits relevant to the farmer with phylogenetic and geographic relationships. While the study is focused on the situation in Algeria, it is paradigmatical for the effect of agricultural globalisation on traditional agricultural systems.
Materials and methods
Plant material
The study included a total number of nine barley accessions comprising landraces and commercial varieties. The seed material was kindly provided by the National Institute of Agronomic Research in Algeria (INRAA) in Constantine. All accessions were six-row barleys and had been collected in 2017. The details of these accessions, their origin, pedigree and morphological characteristics are presented in Table 1.
Table 1. Pedigree, origin and agronomical parameters of the Algerian barley accessions used in the current study

Genotyping
Extraction of DNA
We raised all the accessions to the five-leaf stage in the greenhouse of the Botanical Garden of the Karlsruhe Institute of Technology and used excised leaves shock-frozen in liquid nitrogen. After homogenisation (TissueLyser, Qiagen, Germany), DNA was extracted from aliquots of 60 mg frozen powder using the InvisorbR Spin Plant Mini Kit (Stratec Biomedical AG). We measured quantity and purity of the extracted DNA by spectrophotometry (Nano Drop, Peqlab) and adjusted the DNA concentration to 50–80 μg/μl for amplification by genomic PCR amplification. The samples were stored at −20°C till further analysis.
Amplification by genomic PCR and separation of amplicons by electrophoresis
Ten RAPD markers (Table 2) were selected, during preparatory studies, for their high polymorphism among the tested accessions. Likewise, ten Short Sequence Repeat (SSR) markers (Table 3) were chosen following two criteria: first, to symmetrically represent all chromosomes, and, second, to provide a sufficient degree of polymorphism within the sample set. The two marker sets were used to screen the collection of barley accessions listed in Table 1. We conducted the PCR in a reaction volume of 20 μl containing 1 μg of DNA template, 2 μl of 1x Thermopol Buffer (New England Biolabs), 1 μl of 1 mg/ml bovine serum albumin, 200 mM of dNTPs (New England Biolabs), 0.2 units of Taq polymerase (New England Biolabs) and the respective oligonucleotide primers adjusting with nuclease-free water (Biozym, Lonza) to 20 μl. The concentration of forward and reverse primers was 0.4 mM in case of RAPD, and 0.2 mM in case of SSR.
Table 2. Sequences of the oligonucleotide primers used for Random Amplified Polymorphic DNA, their predicted T m, total number of bands (TB) and number of polymorphic bands (PB) for each marker, along with the calculated polymorphism information content (PIC)

Table 3. Characteristics for the forward (fw) and reverse (rev) oligonucleotide primers used to amplify Simple Sequence Repeats (SSR), (Chr) chromosome location of the respective SSR locus, total number of bands (TB) and number of polymorphic bands (PB) for each marker, along with the calculated polymorphism information content (PIC)

The RAPD analysis was performed with an initial denaturation at 94°C for 5 min, followed by 45 cycles of initial denaturation at 94°C for 1 min, annealing at 36°C for 1 min, and extension at 68°C for 2 min, adding a final extension step of 5 min at 68°C. In case of SSR markers, the initial denaturation was followed by 30 cycles at 94°C for 1 min, annealing for 1 min between 52 and 60°C (depending on the respective primer pair) for 1 min, extension at 68°C for 1 min; and terminal extension at 68°C for5 min. Each PCR amplification was repeated three time to validate the patterns.
We separated the RAPD products by electrophoresis in a 1% agarose gel with 5% v/v SYBR Safe (Invitrogen, Thermo Fisher Scientific, Germany) at a voltage of 100 V, and visualisation by excitation with blue light. A 100-bp DNA ladder (NEB) served to estimate the size of DNA fragments. To achieve a better resolution for the amplicons from the SSR markers, we increased the agarose content of the gel to 2% w/v and allowed the electrophoresis to proceed for 45 min at 130 V.
Data analysis
Amplified fragments of a given size were scored either as present (represented by 1) or absent (represented by 0). The banding pattern was, thus, transformed into binary matrix which was the base of the subsequent analysis.
A pairwise similarity matrix was generated for the RAPD and SSR markers. The similarity table (Table 4) based on the Jaccard indices were computed by the Past software (Hammer et al., Reference Hammer, Harper and Ryan2001) version 3.22.
Table 4. Jaccard coefficients reporting similarity between nine Algerian barley varieties based on the RAPD markers and SSR markers given in Tables 2 and 3 respectively

The polymorphism information content (PIC) was calculated for each marker according to the method of Smith et al. (Smith et al., Reference Smith, Chin, Shu, Smith, Wall, Senior, Mitchell, Kresovich and Ziegle1997), as follows
Where P ij is the frequency of a particular band j, obtained from either the SSR or the RAPD patterns for marker i, and n the entire sum of bands.
The phylogenetic trees were inferred using the Neighbour-Joining algorithm using the software MEGA7 (Version 7.0.14, https://www.megasoftware.net/), the resulting trees were visualised using the integrated Tree Explorer.
A PCA based on agro-morphological traits of barley varieties was conducted using SPSS (version 22.0).
Results
For both, the RAPD and the SSR markers, we obtained well reproducible patterns. For the RAPD markers, we were able to observe in total 50 different bands, 39 of which were polymorphic (Table 2). The number of bands ranged from three to six per marker. The values for Polymorphic Information Content (PIC) were between 0.42 and 0.82, with the highest value scored by the marker BY14. For the SSR analysis (Table 3), the total number of bands was 24, whereby almost all (22) were polymorphic with two to four bands per marker. The only exception was HV13GEIII which was not polymorphic. The most informative marker was Bmag13 (PIC value 0.6), followed by MGB 402 (PIC value 0.59) and Ebmac624 (PIC value 0.5).
SSR-based phylogeny of Algerian varieties correlates well with pedigree
We constructed a binary matrix based on presence or absence of the scored bands. This matrix allowed then to compute the Jaccard coefficients (similarity matrix) by the Past.3 software, for both, the RAPD and the SSR (Table 4), markers. The SSR markers were selected such that they represented all seven chromosomes of the barley genome (Table 3). The similarity for RAPDs (Table 4) ranged between 0.366 (Fouara versus Rihane03) to 0.80 (Fouara and Oued- Athmania), with a mean of 0.677. For the SSRs (Table 4), the values were between 0.25 (Oued- Athmania versus Lamari) up to 0.80 (Saïda183 versus Tichedrett) with an average value of 0.49. Since the SSR markers had been selected for equal representation of all chromosomes, while the RAPD markers are based on unknown locations and, therefore, might represent subject to sampling bias, we plotted the Jaccard coefficients for each pair of genotypes, either using the similarity based on the RAPD markers (online Supplementary Fig. S1a) or that based on the SSR markers (online Supplementary Fig. S1b) as independent variable. Independently of this choice, there was no correlation between the Jaccard coefficients (r 2 = 0.0761, in both cases). This indicates that the distribution of the RAPD markers over the genome differed significantly from the equal distribution for the sampling of the SSR markers and means as well that any conclusions based on these RAPD markers must be taken with care, because these markers are subject to considerable sampling bias. On the other hand, it is coarse graining by asymmetric representation that amplifies even small differences between accessions and, thus, complements the results obtained with the more symmetrically distributed SSR markers.
As to be expected, the phylogenetic trees inferred from the distance matrices based on the UPGMA algorithm differed considerably, depending on which marker type they were based (Fig. 1).
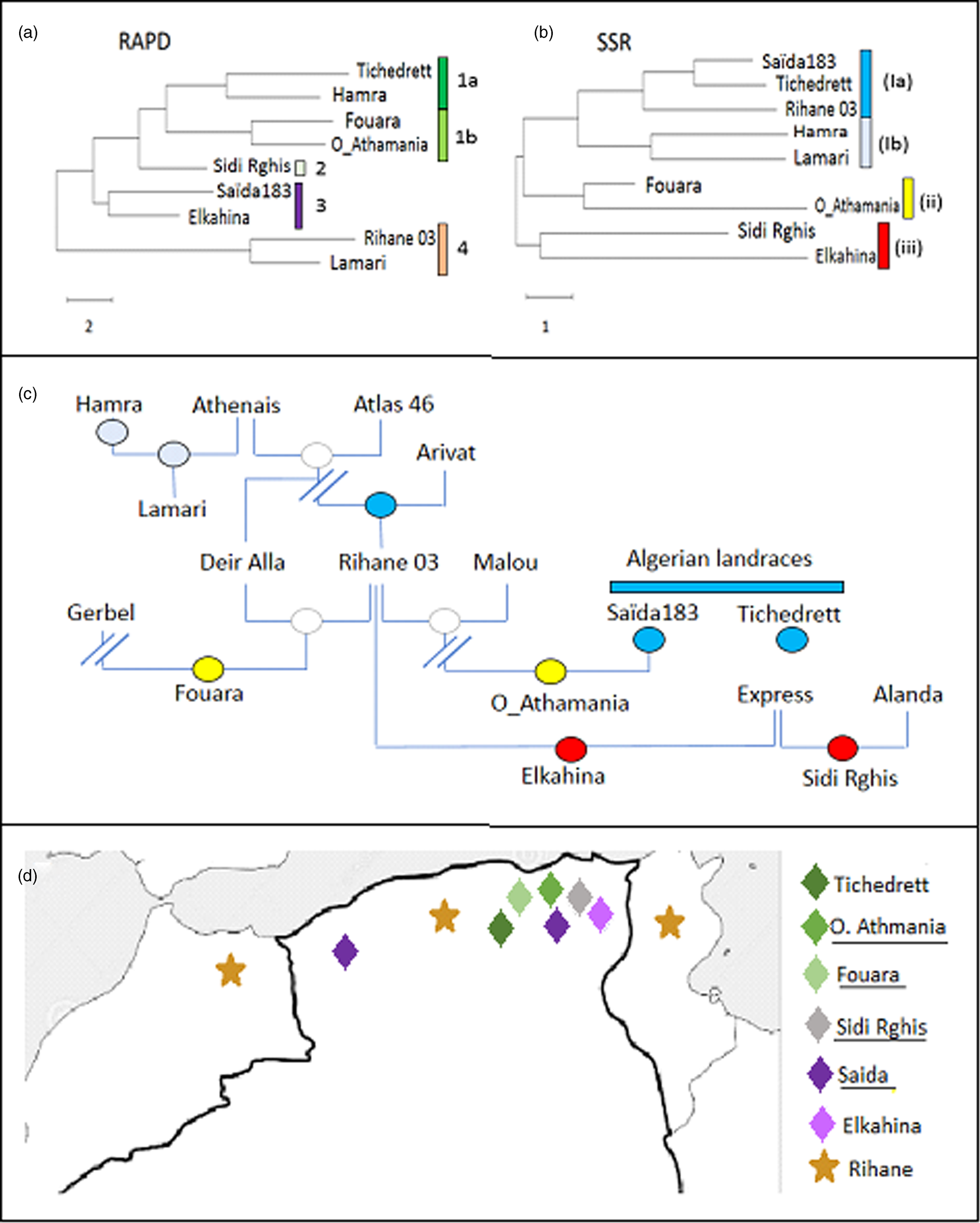
Figure 1. Phylogenetic tree for Algerian barley varieties inferred by Neighbour Joining based on ten RAPD (a), or on ten SSR (b) markers, respectively compared to the pedigree (c). Colour code in the pedigree refer to the clades defined by the SSR markers in (b). (d) Distribution map of the Algerian barley varieties with known geographic origin. Colour code refer to the clades defined by the RAPD markers in (a).
For the RAPDs, three clades can be inferred. The first group contains Lamari and Rihane 03 and is distant from all other accessions. The second clade contains Saïda183 and Elkahina, while the third group consists of two subclades with two relatively close sisters (Fouara and Oued-Athmania; Tichedrett and Hamra), and the basal accession Sidi Rghis.
Likewise, for the SSRs, three clades emerge. However, these are of completely different composition: a first and distant cluster includes Sidi Rghis and Elkahina. A second clade consists of Fouara and Oued-Athmania (that were siblings also in the RAPD tree). The third clade is more complex and consist of two pairs (Hamra and Lamari; Saïda183 and Tichedrett), and Rihane03 located at the base of the Saïda183 / Tichedrett twins.
RAPD markers reflect geographical origin.
The RAPD-based phylogeny seemed to be unrelated to the pedigree (compare Fig. 1(a) and (c)). Here, the varieties Fouara, Oued-Athmania, Sidi Rghis and Tichedrett form a common clade (1). They are all originating from North-Eastern Algeria, meaning that these four accessions cluster with relation to geographical origins as shown in Fig. 1d. For instance, Fouara was first cultivated in the community of Sétif, while Oued-Athmania originates in the city of Oued Athmania, and Sidi Rghis derives from the mountains of the city Oum Bouaghi. Likewise, the origin of, Tichedrett is Sétif as well, while Saïda183 comes from the city Saïda, located in North-West Algeria. Thus, the RAPD markers reflect the geographical origin of theses traditional varieties.
An interesting detail is the close proximity of Saïda183 with Elkahina in clade 3 (Fig. 1a). While the geographical origin of Elkahina is not known, it clearly clusters with the North-Eastern varieties and is separated from the North Western varieties. The name, Elkahina, refers to the ancient berber warrior queen, Dihya Elkahina, linked to the city of Batna in the North Eastern mountains of Algeria. If Elkahina is originating from the North East, the proximity with Saïda183 from the North West in the RAPD-based tree needs an explanation. However, Saïda183 has become the most popular traditional variety and grown all over the Algerian High Plateau, its distribution has shifted to the main cereal-producing areas of Algeria, bordering the Tellian Atlas, where the North Eastern varieties, including Elkahina, originate. Thus, origin of Saïda183 differs from its current geographic focus (represented in the map in Fig. 1d by two locations). it's the clustering of Saïda183 with Elkahina seems to reflect the close proximity between Elkahina and the current area for Saïda183 (rather than its traditional region of origin).
SSR markers reflect phylogenetic origin
In contrast to the RAPD-based tree, the SSR-based phylogeny was significantly linked with the pedigree (compare Fig. 1(b) and (c)). For instance, Sidi Rghis and Elkahina grouped together in clade (iii), and both derive from crosses with the variety ‘Express’. Likewise, Lamari, which derived from a cross involving Hamra, forms a clade with its parent, as it should be expected. Fouara and Oued-Athmania were in one clade, one of the few cases, where the topology was matching that seen for the RAPD-based phylogeny. Both varieties derive from the Syrian variety Rihane 03, which is distantly linked with the Algerian landraces Saïda183 and Tichedrett.
Most importantly, the SSR tree clearly defines Saida183 and Tichedrett as twins (as already seen from a Jaccard score of 80% for this pair, which was the highest value of the whole set (Table 4). This pattern is consistent with the fact that these two varieties are ancient Algerian landraces, with a history dating back to the time of French colonisation. Both are late varieties, tolerant to cold and drought, with very good forage yield, and a high biomass in spring. However, they are sensitive to lodging and fungal diseases, especially Powdery Mildew.
Principal component analysis of phenotypic markers matches to SSR-derived phylogeny
As next step, we assessed, to what degree the genetic relationships are reflected in phenotypic similarity. The seven quantitative traits from the nine barley genotypes were subjected to a PCA using the SPSS (version 9.2) software. The bulk of variation (85.4%) could be explained by the first two principal components (Fig. 2). Principal Component 1 (PC1) contributed with 51.9% to the total variation and was determined by grain height (Eigen value 0.989), time till heading (Eigen value 0.981), time till maturity (Eigen value 0.855), and number of spikes per m2 (Eigen value 0.811). Principal Component 2 (PC2) contributed with 33.47% to the total variation and was mainly brought about by the traits number of kernels/spike (Eigen value 0.892), Total Kernel Weight (Eigen value 0.879), and grain yield (Eigen value 0.814).

Figure 2. Principal component analysis (PCA) for the Algerian barley varieties analysed in this study based on seven agro-morphological traits. 1. Height (cm), 2. Time to heading (days), 3. Time to maturity (days), 4. Number of spikes/m², 5. Number of kernels/spike, 6. Thousand Kernel Weight (TKW), 7. Grain yield. For the colour code refer to the phylogenetic clades inferred from the SSR markers (Fig 1b).
The PCA analysis places the autochthonous Algerian varieties Saïda183 and Tichedrett into a well delineated group positioned in the positive range of PC1 indicating that they mainly differ in terms of height, time to heading, time to maturity and number of spikes. Rihane03 forms an outgroup with a very positive value for PC2, indicating that it differs in number of kernels per spike, grain yield and total kernel weight. The rest of the varieties show strongly negative values for PC1, at high variability of PC2. It is noteworthy that Oued-Athmania, which derives from a cross between Rihane03 and Saïda183, positions roughly in the middle between its parents, right in the centre, where both components are close to 0.
Thus, the phenotypical clustering by PCA is well congruent with the genotypic clusters emerging from the analysis of SSR markers. The autochthonous varieties Saïda183 and Tichedrett are clearly different from the other varieties. Likewise, the grouping of Sidi Rghis, Elkahina, Oued-Athmania and Fouara in the PCA corresponds well to their position in the SSR-derived phylogeny.
Discussion
Elite genotypes have been improved for high yield, which often went along with a loss of resilience factors that are often still present in landraces that had been neglected due to their lower yield. Introgression of alleles that improve the resilience of high-yielding genotypes has become a major strategy in crop breeding and represents a key stone for a more sustainable agriculture in the future. The introgression of qualitative traits has been advanced by the availability of molecular markers (Kumar et al., Reference Kumar, Verma, Singh, Kumar Sharma and Devi2020) and also promotes the improvement of quantitative traits including yield parameters.
As a contribution to this goal, we have analysed the genetic and the phenotypic relationships within a set of nine barley varieties commonly used in Algeria, including the autochthonous landraces Saïda183 and Tichedrett. As tool to assess genetic relationship we used either RAPD or SSR markers that were selected to represent all chromosomes. The genetic data were compared to a set of seven phenotypic parameters as assessed by PCA. We detected a good match of the phenotypic clusters with those emerging from SSR based phylogeny.
In the following, we will discuss first the methodological aspects of our studies before addressing the phylogenetic position of the two autochthonous landraces from Algeria.
RAPD vs SSR based phylogeny as complementary approaches
The use of arbitrary primer pairs to obtain genotype-specific patterns has been a widespread approach because it is versatile and does not require any pre-formed hypothesis on the target sequence of the respective organism. When Williams et al. (Reference Williams, Kubelik, Livak, Rafalski and Tingey1990) introduced this strategy and coined it as RAPD, they demonstrated that the very same primer pairs can be used to discern genotypes of plants, as well as fungi or animals. This makes RAPD a very powerful strategy, if one is addressing organisms, where sequence information is incomplete or even lacking.
Therefore, the RAPDs are still largely used and actual marker of choice in many studies addressing the diversity in plants (Bοusba et al., Reference Bοusba, Gueraiche, Kanouni, Bounar, Djekoune, Khammar and Ykhlef2020; Singh et al., Reference Singh, Kudesia and Bhadauria2020; Somana et al., Reference Somana, Saha, Rashid and Haque2021) (Javed et al., Reference Javed, Ghafoor and Rabbani2022) as well as in epidemiological studies in medical field to better understand the pathogenesis of infections (Stefańska et al., Reference Stefańska, Kwiecień, Górzyńska, Sałamaszyńska-Guz and Rzewuska2022). Considering barley, next to their efficiency to determine the genetic divergence (Fadel et al., Reference Fadel, Abdulhamed and Yousif2022), RAPDs allowed introducing markers linked to different type of stress tolerance, to salt tolerance in cultivated and wild barley (Pakniyat et al., Reference Pakniyat, Namayandeh and Forster2004), and potential primers to identify drought resistant cultivar studied by Nazari and Pakniyat (Reference Nazari and Pakniyat2008). Similar results are reviewed by Younis et al., (Reference Younis, Ramzan, Ramzan, Zulfiqar, Ahsan and Lim2020)
However, the fact that RAPD is completely uncoupled from knowledge about the target sequence, is also its main drawback. In most cases, the sequences behind the banding patterns are not known, nor is it known, how they are distributed over the genome. Moreover, the versatility of RAPD markers comes with a cost – this cost is a relatively low annealing temperature rendering the resulting patterns error-prone limiting their value for phylogenetic studies (van de Zande and Bijlsma, Reference van de Zande and Bijlsma1995). Thus, working with RAPD requires rigorous standardisation of the process and poses high demands on the precision of the PCR and clear definition of the conditions used for electrophoresis. One approach to reduce experimental noise in the use of arbitrary primers are electrophoretic systems, where the conditions of the run are adjusted based on the readout from fluorescently labelled bands (Waller et al., Reference Waller, Furuya and Nick2002). However, such systems are technically demanding and were not available for the current study. To improve the reliability of the RAPD approach, we took utmost care on the purity of the template DNA by using a column-based purification system, and added BSA to the reaction mix, which helps to render the PCR more reliable even under challenging conditions, such as the presence of phenolic compounds that are very common in plants (Jürges et al., Reference Jürges, Sahi, Rodriguez, Reich, Bhamra, Howard, Slater and Nick2018). In addition, we conducted each amplification in biological triplicates to discriminate between informative and accidental amplicons. Despite these precautions one must consider that RAPD markers are not necessarily reflecting different chromosomes to the same degree, such that sampling bias might play a role. To achieve symmetric coverage, one would need a very high number of informative primer pairs, certainly in the range of at least one order of magnitude higher than used in most studies of this kind, including the current investigation.
Considering these limitations of the RAPD strategy, we decided to probe our samples by a second, independent approach, based on SSR markers (microsatellites). These markers can be assigned to a specific locus, and they are inherited in a co-dominant fashion, such that progenies can be directly inferred from their distribution. To identify such markers, original libraries were screened, which was cumbersome and expensive (for review see Squirrell et al., Reference Squirrell, Hollingsworth, Woodhead, Russell, Lowe, Gibby and Powell2003). In the meantime, the availability of genomic data for many species along with the coverage by Expressed Sequence Tags (ESTs) has fuelled the development of SSR marker sets for many crop plants, including barley (Thiel et al., Reference Thiel, Michalek, Varshney and Graner2003). In many cases, the loci of these markers are mapped, such that they can be selected such that all chromosomes are symmetrically represented, which reduces the sampling bias that can distort the pattern obtained with RAPD markers. However, one has to keep in mind that microsatellites are not as neutral as often thought – their evolution is complex and inheritance patterns can be masked by mechanisms such as base slippage or template repair (for review see Ellegren, Reference Ellegren2004).
When we compared the two marker strategies in our sample set, we found that the SSR markers picked up more variation between the varieties (Jaccard index 0.49) as compared to the RAPDs (Jaccard index 0.60). This was also reflected in a higher percentage of polymorphic bands (SSR markers: >90%; RAPD markers: 78%). Our results are well in line with earlier studies, where the two methods had been compared in barley side by side and where the resolution of SSR markers was reported substantially higher than that of RAPD markers (Kraic et al., Reference Kraic, Žáková and Gregová1998). However, this does not necessarily need to be the case – for instance, a comparative study of the two strategies in Citrus rootstocks (Lamine and Mliki, Reference Lamine and Mliki2015) did not find any surplus in resolution by SSR markers over that obtained for RAPD markers, and a study in inbred popcorn lines arrived at a similar conclusion (Leal et al., Reference Leal, Mangolin, do Amaral, Gonçalves, Scapim, Mott, Eloi, Cordovés and da Silva2010).
However, what matters more than these differences in information content is the fact that the clustering inferred from the RAPD markers differed substantially from that seen for the SSR markers and only poorly reflected the pedigree of these accessions. A possible explanation might be sampling bias playing a role, since the genetic position of the RAPD markers is not known, such that asymmetries in their distribution over the chromosomes would go unnoticed. In contrast, the SSR markers were selected such that all chromosomes were represented.
Although the RAPD did not give much significant information in correlation to pedigree or the other studied characteristics, we find that they reflect the geographic relationships between the varieties (Fig. 1d). A closer link between RAPDs and geographic origin was also reported in a study where the two strategies were administered to landraces of pumpkins in South Africa (Ntuli et al., Reference Ntuli, Tongoona and Zobolo2015). In contrast, a study on barley landrace in Tunisia found only poor correlations between RAPD markers and geographic origin (Abdellaoui et al., Reference Abdellaoui, Kadri, Ben Naceur and Bettaib Ben Kaab2010), while during a comprehensive study extending all over North Africa a RAPD-based phylogeny detected two clades with landraces from Tunisia and Algeria grouping distinctly from those originating in Egypt (Allel et al., Reference Allel, Ben-Amar, Lamine and Abdelly2017).
What might be the reason for this geographic representation between traditional varieties and the phylogeny inferred from RAPD markers? Traditional varieties are usually cultivated as landraces, meaning that a part of the harvest is retained and resown the following year. In contrast to modern cultivars that are genetically homogenous and standardised by the registration process, landraces represent populations that adjust to the conditions of a particular region and also entertain gene flow with neighbouring landraces, often by human exchange, such as trading. Thus, there is a correlation between genetic distance and geographic distance. Since RAPDs are selected with respect to polymorphism, they highlight such differences between neighbouring landraces to an extent that would go unnoticed with SSR markers picked for the symmetric representation of chromosomes. In other words: it is the sampling bias of RAPDs that ‘coarse-grains’ the subtle differences deriving from geography.
Therefore, RAPD markers are more than a poor and preliminary alternative to SSR markers, but actually a complementary approach. Their value should become manifest mainly in the case of autochthonous varieties, while this link with geography would be expected to erode in modern varieties with their often hybrid or complex pedigree, as well as in varieties that become popular beyond their region of interest (Serpoush et al., Reference Serpoush, Garcia-Cozar, Salayeva and Ojaghi2022). The case of Saïda183, which has shifted from its ancestral region in West Algeria to the East of the country gives evidence for this conclusion.
Thus, we propose to analyse traditional varieties not only with SSR markers, but also to include a RAPD-based phylogeny as complementary strategy. However, we conclude that the value of RAPDs will diminish, when it comes to modern cultivars or to varieties that have already undergone a certain degree of commercialisation.
To what extent a given arbitrary marker (such as RAPD or SSR) reflects phylogenetic relationships, depends also on the question, to what extent it is coupled to morpho-agronomic traits. To characterise genetic resources with respect to their morpho-agronomic traits is fundamental, therefore (Zeng, Reference Zeng2015). In our set of genotypes, we detected a positive and significant correlation between days to heading, height, days to maturity and number of spikes per area, while days to heading correlated negatively with the number of grains per spike. Numbers of grain per spike has been suggested as proxy for 1000-grain weight (Al-tabbal and Al-Fraihat, Reference Al-tabbal and Al-Fraihat2011), but findings where there is a high, but negative correlation, challenge this suggestion (Rahal-Bouziane et al., Reference Rahal-Bouziane, Merdas, Merzoug and Abdelguerfi2015). In congruence with the findings on other barley landraces (Setotaw et al., Reference Setotaw, Dias and Missio2010; Drikvand et al., Reference Drikvand, Salahvarzi, Salahvarzi and Hossinpour2012), we observe that parameters related to the extent (plant height) or duration (days to heading) of the vegetative phase are the main drivers of phenotypic variability.
Based on the PCA of the phenotypic parameters, the landraces Saïda183 and Tichedrett are clearly delineated as a separate group, as they require the longest time till heading and maturity, in line with findings of Ben Naceur et al. (Reference Ben Naceur, Chaabane, El-Faleh, Abdelly, Ramla, Nada, Sakr and Ben Naceur2012) on Saïda183. With respect to grain yield and numbers of grains per spike (comprised by the second principal component), Rihane03 excels, which is in good agreement with the results by Reguieg et al. (Reference Reguieg, Labdi, Benbelkacem, Hamou, Maatougui, Grando and Ceccarelli2013), where this genotype generated the highest yield in the hands of four scored farmers. Thus, these three varieties stick out from the entire set and render them favourable for the use as fodder, which is the economically most relevant use in Algeria.
On the other hand, the remaining varieties provide certain advantages as well, such as earlier heading, higher productivity or better resistance to diseases. Mostly Fouara and Hamra (Barberousse) have been studied in this respect. For instance, Fouara not only performed well with respect yield (Taibi et al., Reference Taibi, Mahdad and Gaouar2016), but also can cope with Helinthosporiosis, Anthrax and Rhyncosporiosis (Silem and Tioub, Reference Silem and Tioub2017). In contrast, the other varieties (Elkahina, Sidi Rghis, Oued-Athmania and Lamari) have not been addressed prior to the current study.
The phenotyping clustering obtained by the PCA is matching the phylogeny obtained by SSR markers closer than that for RAPD markers. This does not necessarily need to be the case – if the RAPD markers would be located close to the genetic factors driving the phenotypic variation, they would reflect agro-morphological traits more truly. However, since their selection is arbitrary, one needs to be very lucky to select those that are linked with a trait. Thus, a strategy, where the markers are deliberately chosen such that different locations of the genome are more or less symmetrically represented, is more promising, if one does not have any a-priori knowledge on genetic linkage with a marker and a trait of interest.
Molecular phylogeny support autochthony of Tichedrett and Saïda183
Barley production in Algeria is dominated by mixed cultivation of local varieties with genotypes introduced from the Near East (Ullrich, Reference Ullrich2011). In addition, a small number of landraces are in use that are used beyond a particular location, and partially even officially registered. Among these landraces, Saïda183 and Tichedrett are the most popular, and preferred by the farmers. Thus, they are economically efficient despite the fact that they are susceptible to most barley diseases. In Algeria, the selection of the cereals was based on mass selection, promoted by Ducellier as genealogic selection more than 80 years ago (Agria, 2022). Examples for the success of this strategy are the Durum wheat varieties BIDI17 or Oued Zenati368. For barley, the varieties Saïda183 and Tichedrett were selected among a large number of local accessions (Reguieg et al., Reference Reguieg, Labdi, Benbelkacem, Hamou, Maatougui, Grando and Ceccarelli2013) and have, since their release in the late 1990s, acquired the largest economic impact, although they are susceptible to most barley diseases. Geographically Saïda183 was developed in Western high plateaus of Algeria and is more adapted to semi-arid conditions, while Tichedrett originates from the Eastern high plateaus and can cope with higher rainfall. Both varieties are also found in Morocco and Tunisia.
Both, Saïda183 and Tichedrett are clearly different from the other barleys in their phenotypical traits (Fig. 2), which is mainly linked with a long time required for heading (a trait that has been selected by the predominant usage as fodder). However, they are also grouped by SSR-based phylogeny (Fig. 1b), which supports their autochthonous genesis in Algeria. Interestingly, the variety Rihane (Fig. 1b) was the closest sister among the tested genotypes. This variety has been propagated through the Near East and North Africa through a programme by ICARDA (Mazid et al., Reference Mazid, Aw-Hassan and Salahieh1995), and has a complex pedigree (Fig. 1c) deriving from parental lines that are rooted in landraces from Germany, but also the Greek landrace Athenais. Barley had been cultivated in the Maghreb since at least 2000 years, as it is already mentioned in the agricultural scripts of Mago of Carthago. Moreover, barley was found in finds dating back 2500 years ago (van der Veen et al., Reference van der Veen, van Zeist and Bottema2001), and was either used for beer or as fodder. As Saïda183 and Tichedrett, it was a six-rowed barley. Whether the Phenicians that had founded Carthago, brought with them from barley from the Near East, or whether it had reached Algeria even earlier, is not known and will not be clarified by archaeology, nor fragmentaric historic documents. However, the SSR data indicate that both landraces are autochthonous with a long local history in Algeria.
Conclusion and outlook
To safeguard food security under the pressure of actual global climate change requires introgression of resilience factors into the high-yielding crops dominating current agriculture. Traditional varieties have often adapted to the challenges of a particular region or the requirements of a particular form of usage and have experienced a renaissance as genetic resources. Characterisation of traditional barley varieties from North Africa is promising with respect to detect traits linked with resilience to drought. Using varieties from Algeria as paradigm, we have used two widespread approaches, RAPD and SSR, to infer the phylogenetic relationship between varieties commonly used in Algeria. The SSR markers were selected such that the different chromosomes were represented symmetrically, and the resulting phylogeny was in good congruence with the pedigree. In contrast, the RAPD-based phylogeny did not the pedigree at all, which might be due to sampling bias with respect to the distribution of these markers across the genome. On the other hand, the RAPD markers related to the geographic origin of the accessions. Agro-phenotypic parameters, as assessed by PCA, along with the SSR-based phylogeny, supported the autochthonous origin of the two relevant landraces Saïda183 and Tichedrett. Both varieties represent interesting candidates for genetic introgression and show a more distant relationship to the variety Rihane, originating from the Near East. Since Saïda183 and Tichedrett, as well as Rihane are also grown in other Maghreb countries, it would be highly interesting to compare the diversity of these varieties and landraces between Algeria, Tunisia and Morocco.
To get further insight into the history of evolution and migration of Saïda183 and Tichedrett, we are currently addressing their relationship with a broader set of traditional varieties from the Near East and Europe.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262123000291
Acknowledgements
We thank the members of the Botanical Institute of the Karlsruhe Institute of Technology: Dr Daniela Rios and Dr Sascha Wetters for introduction into molecular methodology, and Dr Adnan Kanbar for providing RAPD markers.
Financial support
This research did not receive any specific grant from funding agencies in the public, commercial, or not –for- profit sectors.










