To the Editor—Clostridium difficile is a frequent cause of antibiotic-associated diarrhea and healthcare-associated infections.Reference Bartlett 1 , Reference Magill, Edwards and Bamberg 2 Epidemics of C. difficile infection (CDI) have occurred in North America and Europe over recent decades. In particular, epidemic C. difficile (sequence type [ST] 1/027/NAP1) has rapidly emerged in the past decade as the leading cause of C. difficile–associated diarrhea worldwide, resulting in high morbidity and mortality in hospitalized patients. Very little is known about the epidemiology of C. difficile–associated diarrhea outside of North America and Europe. However, the hypertoxigenic strain C. difficile RT027 has rarely been detected in Asia to date, specifically not in China before 2013.Reference Wang, Zhou and Wang 3 In addition, cases of C. difficile RT078 have not been reported in China. We report a novel binary toxin–positive non-027, non-078 C. difficile associated with severe diarrhea recently identified in China.
A 65-year-old man was admitted to Xiangya Hospital in Changsha, China, with fever, headache, diarrhea, and impaired consciousness. He had been diagnosed with a central nervous system infection 9 days earlier in a local hospital, presumptively diagnosed as suppurative or tubercular meningitis. Antimicrobial therapy was initiated with ceftriaxone over the next 4 days in that local hospital. The patient’s condition did not improve; he developed a pulmonary infection, with Acinetobacter baumannii isolated from the sputum, for which he was intravenously treated with cefepime for 5 days. The consciousness level of the patient deteriorated, and he developed severe diarrhea and experienced bouts of vomiting. Therefore, on February 8, 2014, he was transferred to Xiangya Hospital. Stool specimens collected during hospitalization tested positive for C. difficile toxin B by polymerase chain reaction assay, and C. difficile was confirmed by culture and biochemical characteristics. Enteral vancomycin was prescribed for the patient, but symptoms further deteriorated and he died on day 4 of hospitalization.
Stool specimens from the patient were cultured for anaerobic bacteria, and C. difficile was isolated and identified. The toxin genes tcdA, tcdB, cdtA, and cdtB were detected by polymerase chain reaction assay.Reference Pituch, Kreft and Obuch-Woszczatynski 4 The isolate (LC693) was positive for toxin A, toxin B, and binary toxin. DNA sequence analysis of the toxin gene to the genome sequence of CD630 (GenBank accession No. AM180355.1) revealed an 18–base pair deletion (nucleotides 330-347) located in tcdC (682 base pairs, GenBank accession No. KM609431.1) of LC693. It did not express a variation at nucleotide position (nt) 117 (associated with RT027). However, there was a point mutation at nt 184, resulting in the introduction of a premature stop codon (TAA) in the putative TcdC protein (Figure 1). Multilocus sequence typing indicated the isolate was ST201. Alleles for the profile were adk-1, atpA-6, dx-4, glyA-7, recA-2, sod-8, and tpi-31, making the Xiangya hospital clinical isolate LC693 unique from the RT027 (ST1, R20291, GenBank accession No. FN545816.1) and RT078 (ST11, M120, GenBank accession No. FN665653.1) clones. Whole genome sequencing of C. difficile LC693 was performed using a MiSeq (Illumina) by PE300 strategy. Approximately 424 Mb clean data were obtained, with a mean read length of 300 base pairs, 100 times coverage of the approximately 4.07 Mb genome. hylogenomic analysis showed that the LC693 was more closely related to the RT027 (CD196, R20291) cluster than to the RT078 (M120) (Figure 2). Between LC693 and R20291, the single-nucleotide polymorphisms were 35,505; between LC693 and M120, the single-nucleotide polymorphisms were 87,672.

FIGURE 1 Comparison of the tcdC nucleotide sequences of the Clostridium difficile reference strain CD630 with the isolated strain from a patient with diarrhea that contains both the 18–base pair deletion (from nucleotide position [nt] 330–347) and a premature stop codon TAA (from nt 184–186); Query, tcdC gene (from nt 1-681) of the isolated strain LC693; Subject (Subjct), tcdC gene of the reference strain CD630 (from nt 805008–804310).
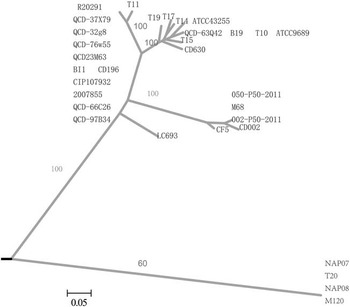
FIGURE 2 Whole-genome phylogenomic tree of strains LC693 (sequence type [ST] 201) compared with 31 publicly available Clostridium difficile genomes. Bootstrap values are labeled along main branches. Bar=0.05.
The incidence and associated mortality of CDI have been increasing. This changing epidemiology has coincided with the emergence and rapid spread of C. difficile RT027/078, involved in several large outbreaks of severe CDI in North America and Europe. Furthermore, hypervirulent RT027 and RT078 have been associated with more severe disease because of the production of higher amounts of toxin A and B due to tcdC deletion.Reference Carter, Douce and Govind 5 , Reference Goorhuis, Bakker and Corver 6 Sequence analysis of tcdC of the isolate LC693 showed an 18–base pair deletion and a premature stop codon (TAA), compatible with the genotype of a hypertoxigenic strain. However, it also differed from RT027 and RT078, indicating possible existence of a new hypervirulent strain in China. Our findings were similar to those of Lim et al,Reference Lim, Stuart and Mackin 7 who identified and characterized a C. difficile strain associated with a severe clinical phenotype that genetic analysis showed to be ST41/RT244 that was also different from RT027 and RT078 in Melbourne, Australia. There were 10,803 single-nucleotide polymorphisms between ST41/RT244 and RT027. Further tests are required to determine the ribotype and toxin production of the isolate LC693, and study of the epidemiology of the strain will be helpful in monitoring its spread in future. However, laboratory confirmation for C. difficile is not routinely performed in hospitals in China, so there is not a current CDI surveillance system or plan in China. Many C. difficile–associated diarrhea cases are suspected by a subjective judgment and clinical assessment of patient characteristics in most hospitals. Such cases are administered empirical treatment. Estimating the incidence of CDI in China is difficult because there are no national surveillance data. In order to enable a more accurate diagnosis of CDI and to prevent outbreaks of such infection, implementing standardized laboratory-confirmed diagnosis and treatment procedures and developing healthcare-associated infection surveillance programs of C. difficile should be encouraged in China.
NUCLEOTIDE SEQUENCE ACCESSION NUMBER
The tcdC nucleotide sequences of the isolate LC693 was submitted to GenBank. The sequence is available under accession No. KM609431.1.
Acknowledgments
We thank Carol Y. Rao, John D. Klena, Yuan Li, Yang Huai, Yuzhi Zhang, and Hongbing Jia for technical support and language proofreading.
Financial support. US Centers for Disease Control and Prevention (Cooperative Agreement No. 5U2GGH000018).
Potential conflicts of interest. All authors report no conflicts of interest relevant to this article.






