Published online by Cambridge University Press: 14 April 2009
Associative overdominance arises at an intrinsically neutral locus through its non-random association with overdominant loci. In finite populations, even if fitness is additive between loci, non-random association will be created by random genetic drift.
The magnitude of such associative overdominance is roughly proportional to the sum of 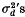 between the neutral and the surrounding over-dominant loci, where
between the neutral and the surrounding over-dominant loci, where 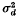 is the squared standard linkage deviation, defined between any two loci by the relation
is the squared standard linkage deviation, defined between any two loci by the relation
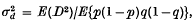
in which p and 1 – p are frequencies of alleles A1 and A2 in the first locus, q and 1 – q are frequencies of alleles B1 and B2 in the second locus, and D is the coefficient of linkage disequilibrium. A theory was developed based on diffusion models which enables us to obtain formulae for 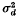 under various conditions, and Monte Carlo experiments were performed to check the validity of those formulae.
under various conditions, and Monte Carlo experiments were performed to check the validity of those formulae.
It was shown that if A1 and A2 are strongly overdominant while B1 and B2 are selectively neutral, we have approximately

provided that 4Nec ≫ 1, where Ne is the effective population size and c is the recombination fraction between the two loci. This approximation formula is also valid between two strongly overdominant as well as weakly overdominant loci, if 4Nec ≫ 1.
The significance of associative overdominance for the maintenance of genetic variability in natural populations was discussed, and it was shown that Nes′, that is, the product between effective population size and the coefficient of associative overdominance, remanis constant with varying Ne, if the total segregational (overdominant) load is kept constant.
The amount of linkage disequilibrium expected due to random drift in experimental populations was also discussed, and it was shown that 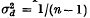 in the first generation, if it is produced by extracting n chromosomes from a large parental population in which D = 0.
in the first generation, if it is produced by extracting n chromosomes from a large parental population in which D = 0.