Hepatitis B virus (HBV) infections lead to a wide spectrum of liver injuries ranging from acute self-limiting infection and fulminant hepatitis to chronic hepatitis, cirrhosis and hepatocarcinoma. Although, HBV infection is predominantly of sporadic nature, epidemics in the past have been reported mainly in healthcare settings highlighting the failure of healthcare personnel to adhere to good medical practice and infection control measures. The USA reported 33 outbreaks of HBV or hepatitis C virus (HCV) from 1998 to 2008 [Reference Thompson1]. In India, an outbreak of HBV with high mortality (7/15, 46·7%) based on serological diagnosis confirming anti-delta positivity in three cases including one survivor with no molecular data was reported in Gujarat in 1997 [Reference Singh2]. Around February–March 2009, one of the most severe outbreaks of HBV infection erupted suddenly around the Modasa township of Gujarat state in India, affecting over 500 people and causing the death of 93 people within just 3–4 weeks [Reference Gandhi3, Reference Arankalle4]. The characteristic of this outbreak was its short incubation period; all patients tested positive for HBsAg with high serum levels of alanine aminotransferase (ALT) and rapid development of fulminant hepatitis leading to death from hepatic and renal failure. Epidemiological investigation traced the source of infection to contaminated syringes and needles, as all dying patients revealed a history of having received therapeutic injections from the same physician, which led to his arrest and the closure of his clinic. Violation of infection control practices became a major news story after the outbreak. Considering the aforesaid details, we investigated this outbreak for the possible link of specific mutations in the basal core promoter (BCP)/precore/core and partial pol genes of the circulating HBV strains with fulminant exacerbation of the disease outbreak.
Since the National Centre for Disease Control (NCDC) was officially designated as the investigating agency, the HBV outbreak investigation did not require any prior ethical clearance. The present study was designed based on the case definition of an acute case according to national guidelines, i.e. the patient should be positive for hepatitis B surface antigen (HBsAg), and have an ALT level of >400 IU. Serum samples from 21 acute fulminant hepatic failure (FHF) cases and 10 acute, self-limiting hepatitis B (ASH) cases were collected and transported to NCDC under cold conditions (i.e. cold packs maintained at 0–4°C). Although the sample size was not adequate, probably due to the high number of deaths causing health officials to focus mainly on curbing mortality, which limited the number of samples they collected. However, with complete outbreak details available, we were able to closely observe the demographic details (Table 1). All 21 FHF patients had a history of receiving injections during the last 6 months at the clinic of the suspected physician. All the serum samples were tested for HBsAg, antibodies to hepatitis B surface antigen (anti-HBs), hepatitis B e antigen (HBeAg), antibody to HBeAg (anti-HBe), anti-HBc (immunoglobulin IgG and IgM), anti-HAV (IgG and IgM), anti-HCV and anti-HEV by micro-particle enzyme immunoassay (Abbott Laboratories, USA) according to the manufacturer's protocol at the central Hepatitis Laboratory of our institute. Viral DNA was extracted from serum samples using a commercial DNA mini kit (Qiagen GmbH, Germany). A nucleotide (nt) sequence of around 1078 bp encoding for precore/core and partial pol genes was amplified with forward primer (5′-GGAGTTGGGGGAGGAGATTA-3′) (nt 1736–1755) and reverse primer (5′-AGGCGCTACGTGTTGTTTCT-3′) (nt 2786–2805) designed with Primer 3 (v. 0.4.0) software (http://fokker.wi.mit.edu/primer3/). Next, 5 μl DNA was used for PCR in a total reaction volume of 25 μl containing 10 pmol of each oligonucleotide in a 1× PCR reaction buffer (final concentration): 2·5 mm MgCl2, 200 μm dATP, dGTP, dCTP, dTTP and 2·5 U AmpliTaq Gold (Applied Biosystems, USA). The thermal cycling parameters included an initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 59°C for 1 min and extension at 72°C for 2 min, followed by a final extension at 72°C for 10 min. The amplified product was purified and sequenced from both directions using fluorescence-labelled dideoxynucleotides on an automated DNA sequencer (ABI 310 Genetic Analyser; Applied Biosystems, USA) using the dideoxy terminator cycle sequencing chemistry following the protocol used by us previously [Reference Chaudhary5]. For sequence alignment and phylogenetic analysis, we selected the GenBank sequences with the best and highest scoring matches with our sequences in a NCBI BLAST search. Multiple sequence alignment was performed using Bioedit version 7.0.4.1 [Reference Hall6]. Genetic distances were calculated using the Kimura two-parameter algorithm and a phylogenetic tree was constructed by the neighbour-joining method. To confirm the reliability of the pairwise comparison and phylogenetic tree analysis, bootstrap resampling and reconstruction were performed 1000 times. Phylogenetic analysis was done using MEGA version 4.1 [Reference Tamura7]. The unpaired t test was used to compare group means of nucleotide sequence divergences. Two-tailed P values of <0·05 were considered statistically significant. Calculations were performed using Stata statistical software (release 5·0; Stata Corporation, USA). The χ2 test was used to compare different attributes in demographic analysis (Table 1). The new sequences obtained in this study were submitted to EMBL and assigned accession numbers JQ287601–JQ287621.
Table 1. Number and percentage of cases and deaths for age group and gender
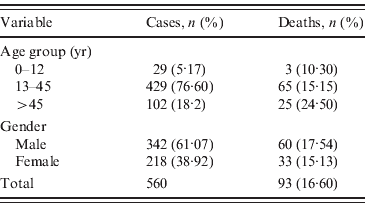
Demographic analysis revealed a total of 560 hepatitis B cases reported with 93 (16·60%) deaths in one of the most lethal outbreaks ever reported in India. Statistically significant difference was observed between 61·07% male and 38·92% female cases reported in this hepatitis B outbreak (P<0·05) (Table 1). The maximum number of cases, i.e. 429 (76·60%) cases with 65 (15·15%) deaths, were reported in the 13–45 years age group, followed by 102 (18·2%) cases with 25 (24·50%) deaths in the >45 years age group, the least number of cases occurred in the 0–12 years age group (29 cases, 5·17%; three deaths, 10·30%). Considering the percentage of deaths in each age group, we observed no significant difference between different age groups (P>0·05). Serological analysis revealed that all 21 FHF samples tested positive for HBsAg and HBc IgM suggesting that all the cases were of acute HBV infection further confirming the aetiology of the outbreak. HBeAg was detected in six cases and found negative in 15 cases. Anti-HBe was detected in 12 cases and both markers were detected in three cases. None of the samples were reported to be positive for HDV or other hepatitis markers (HAV, HCV, HEV), thereby excluding any possibility of co-infection with other hepatitis viruses. Moreover, the level of ALT, an indicator of liver necrosis, was determined. Compared to normal levels of 7-56 IU/l, the average serum ALT level found in the fulminant cases was 2391 IU/l. The serum ALT levels exhibited a marked 46-fold rise, with reference to the upper limit of the normal range. Of the 10 ASH samples, all were found positive for HBsAg and anti-HBc IgM; nine were further positive for HBeAg and one positive for anti-HBe.
Sequencing analysis revealed 19 mis-sense mutations exclusively in the 21 FHF samples and none in the 10 ASH serum samples: two in the BCP region (A1762T, G1764A), 10 in the C region (three in precore and seven in core) which notably included an isoleucine (Ile, ATC) insertion after nt 1843, G1862A and G1896A transversion, and seven in the partial P region, among which two rare mutations were observed at amino-acid positions 89 and 119 (Table 2). Phylogenetic analysis of our sequences with database sequences revealed that all 21 FHF cases belonged to genotype D (subgenotype) (Fig. 1). Analysis of the 10 ASH cases revealed their similarity to wild-type precore/core and polymerase (pol) gene sequences which ruled out the possibility that HBV strains responsible for this outbreak pre-existed in this geographical area. Among these cases eight were found to belong to genotype D and two to genotype A (data not shown).
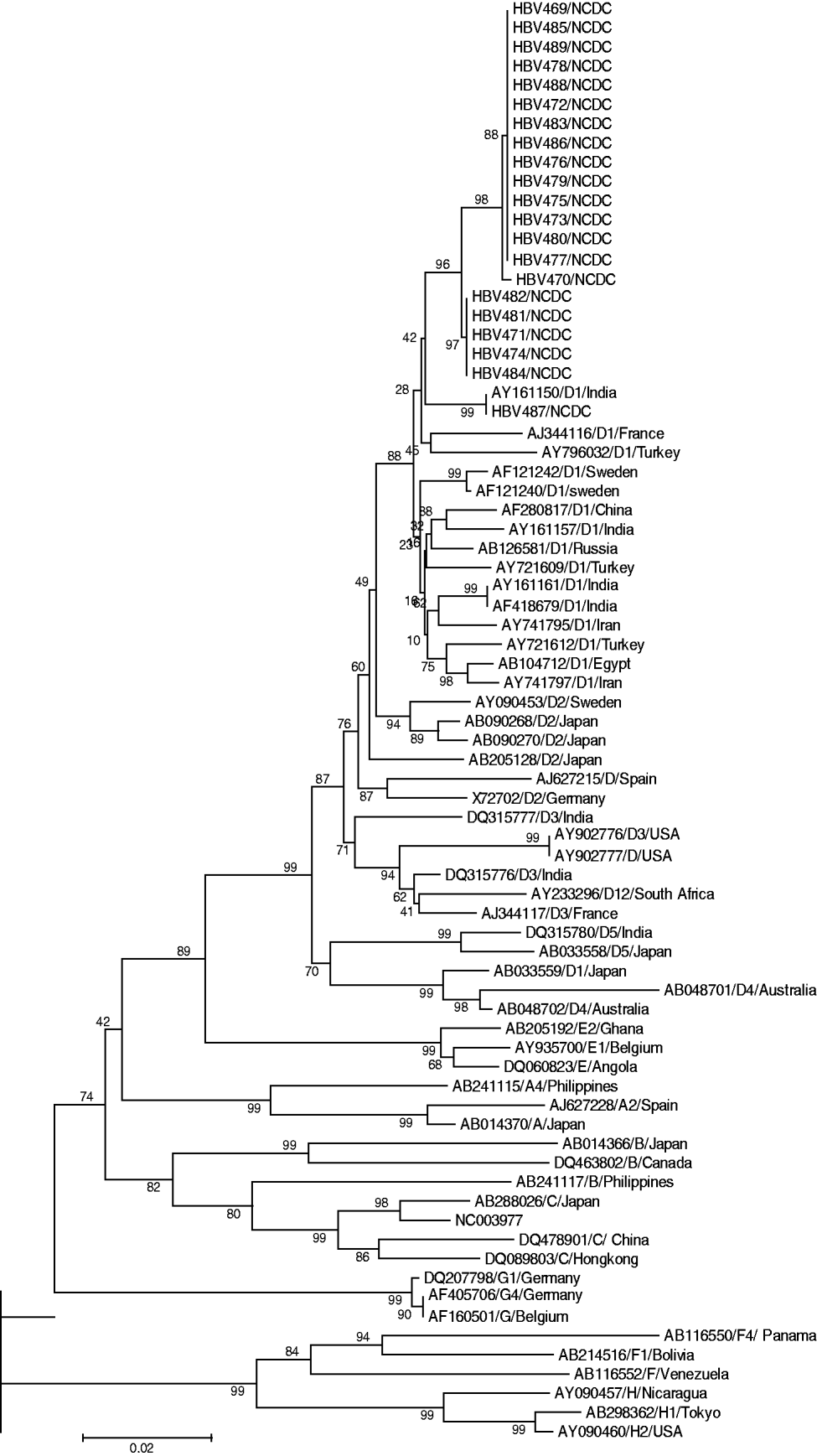
Fig. 1. Phylogenetic analysis constructed using MEGA software version 4.0.2 showing our sequences, with representative sequences of all HBV genotypes identified by respective accession number and genotype, along with bootstrap values.
Table 2. List of mutations in the precore/core and pol genes in the outbreak samples of Gujarat, India
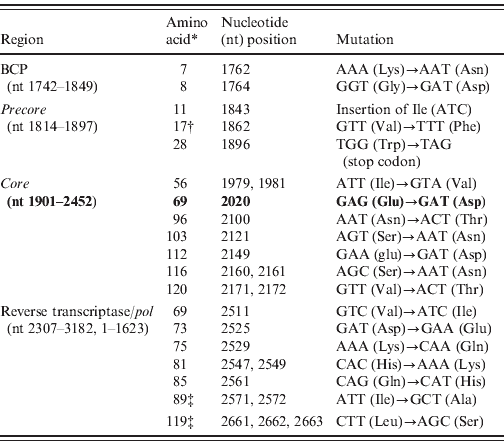
* Amino acid positions are numbered from the start codon of each protein.
† Four strain HM042274, HM042275, HM042279 and HM042280 were observed with this mutation.
‡ Rare mutation.
High mortality during fulminant hepatitis B outbreaks poses a serious threat to a vulnerable community. Since studies on fulminant hepatitis B are usually performed late in the course of the disease, it was difficult to reconstruct the sequence of virological and immunological events that led to mass hepatocellular injury [Reference Mondelli and Eddleston8]. In terms of the percentage acquisition of infection in males and females, a predilection of acquisition in males was observed. This could be attributed to the fact that in Gujarat, a state with poor literacy and a skewed sex ratio, females may not take good care of their own health, resulting in a very small percentage of women attending clinics for their illness. Thus, fewer females were affected in this deadly HBV outbreak which was epidemiologically linked with repeated usage of syringes and needles. In terms of the percentage of deaths, numbers were equally distributed among the sexes. Thus, the female physiology did not affect the severity of the infection. This outbreak was significant for its rapid spread and high mortality which encouraged us to discover the possible cause and source of the outbreak. Although unusual, the outbreak was evidenced by the presence of HBsAg and IgM anti-HBc in the patients. Considering co-infection with HDV as a possible reason for high mortality, all the cases were screened for HDV and other hepatitis viruses such as HAV, HEC or HCV. None of the samples were found positive for co-infection which excluded this as the possible reason for the outbreak. Epidemiological investigation by health officials identified an association of cases with a history of injection and a physician treating very large numbers of patients on the basis that all 40 dying patients gave information about receiving injections from the suspected physician. This physician was later arrested and his clinic closed down following a local court directive. The need for stringent legal action was seen as necessary to deal with such a case of medical negligence which would otherwise have caused further fatalities in the population. This outbreak prompted healthcare authorities to drive the campaign to check for the compliance of local hospitals regarding their infection control practices. Similarly, a study by Samandari et al. found a lack of infection control measures in the USA where a large outbreak of HBV infection was associated with frequent injections from a physician [Reference Samandari9]; while Harpaz et al. implicated a surgeon in the transmission of HBV to 19 patients [Reference Harpaz10]. Phylogenetic analysis of genotype D1 and close sequence homology in all FHF cases strongly indicate the involvement of a single hepatitis B mutant strain/source. Investigating the same outbreak, Arankalle et al. reported the presence of precore and BCP mutants and four amino acid substitutions, genotype D with D1 being present in all FHF cases while D2 was associated with self-limiting acute viral hepatitis cases [Reference Arankalle4]. Expanding on the findings of Arankalle et al. we surmise that BCP mutations at nt 1762 and nt 1764, insertion of Ile after nt 1843, transversion of G to T at nt 1862 (changing the specificity of codon 17 from valine to phenylalanine, responsible for abrogating cleavage of p25 by the cellular signal peptidase), and transition of G to A at nt 1896 led to the creation of a premature stop codon in the ORF of the precore region by truncating the precore/core protein into a 28-amino-acid peptide against the original 29-amino-acid precore and 181-amino-acid core genes. This resulted in inhibition of HBeAg expression through transcriptional down-regulation and enhanced viral replication which implicated all 21 cases to be circulating BCP/precore mutants; and the probable association of these mutant viruses with fulminant hepatitis and mortality, as HBeAg is a known immunomodulator that prevents exceedingly strong cytotoxic immune responses, as suggested in previous studies [Reference Arankalle4, Reference Chandra11–Reference Li13]. This is further supported by serological analysis where the presence of HBeAg in six patients, anti-HBe in 12 and both markers in three patients indicated rapid mutation of the virus to convert into the mutant form which probably took over the initially circulating wild virus. By contrast, none of the reported mutations were found in the self-limiting HBV cases whose sequences exhibited a wild-type pattern. Further, the outbreak isolates were found to exhibit mutations of unknown clinical significance in the precore/core and partial pol genes, which may predict these HBV isolates as mutants divergent from all known DNA sequences; however, the significance of these mutations in the pathogenesis of the fulminant exacerbation of the disease needs to be elucidated [Reference Garmiri14]. All observed mutations in the viral variants possibly account for clinically aggressive and more virulent fulminant HBV disease [Reference Shanmugam15, Reference Liaw and Chu16]. A specific HBV mutation resulting in a stop codon at the precore end region has been associated with a clinically aggressive form of chronic liver disease in a group of Mediterranean patients with anti-HBe and high levels of circulating HBV DNA [Reference Brunetto17]. In addition, we also examined the partial pol gene in which seven mis-sense mutations were found. The biological effect of these mutations is not well understood and their role in producing a mutant HBV strain leading to a more fulminant course of hepatitis B requires further explanation which could be established by site-directed mutagenesis of the representative HBV clone ayw and transfection of HuH-7 cells followed by Southern blot analysis as performed previously [Reference Blum18]. However, we hypothesize that these pol gene mutations might have caused some alteration in either of the four domains from 5′ to 3′, a region coding for the terminal protein, a spacer domain, a DNA pol gene and an RNase H domain to affect encapsidation of pre-genomic RNA to enhance the pathogenicity of the HBV mutant strain in some undefined manner. Our finding of genotype D1 and the clinical presentation of the outbreak supported the notion that the infecting genotype may be associated with the viral load which in turn was correlated with more pronounced liver disease progression, responsible for the high number of deaths within a short period [Reference Vivekanandan19]. In northern India, Thakur et al. found that genotype D was associated with severe liver disease compared to genotype A [Reference Thakur20]. We conclude with the view that the BCP/precore mutant, the mutation in core and partial pol genes along with the finding of genotype D (subgenotype D1), and the clinical presentation of cases in terms of positive HBsAg, elevated ALT indicative of hepatocyte damage, liver necrosis, and enhanced level of anti-HBc cited in the study could corroborate the statement that this outbreak was caused by a mutant HBV strain having high virulence and replication. The finding of this study strongly warrants periodical monitoring of the discharged treated cases to check for virus clearance in order to curb its further spread and strenuous efforts to increase awareness of bloodborne infection among medical practitioners, and the public at risk, to ensure that standard precautions are always practised, and also to restrict the re-use of syringes and needles to prevent this often fatal infection. We believe all medical practitioners found guilty of medical negligence leading to the loss of human life must be deprived of their medical degree with an accompanying legal punishment in order to curb such serious human error.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the financial support from the National Centre for Disease Control (NCDC) during the course of this study.
DECLARATION OF INTEREST
None.





