Introduction
One obvious question that arises in infectious disease, is why do some animals/humans become sick and/or die but others are resistant, survive illness or show no signs or symptoms of disease? This led Casadevall and Pirofski (Reference Casadevall and Pirofski2018) to a consideration of the factors that underly susceptibility to disease and the identification of 11 attributes that determine the outcome of host–microbe interaction. These factors were: Microbiome, Immunity, Sex, Temperature, Environment, Age, Chance, History, Inoculum, Nutrition, and Genetics, the first letters of which spell the acronym MISTEACHING. The MISTEACHING framework has been extensively applied to human (Casadevall and Pirofski, Reference Casadevall and Pirofski2018) but not, to our knowledge, strict veterinary pathogens. The purpose of this review is to demonstrate the utility of the MISTEACHING framework by applying it to Actinobacillus pleuropneumoniae (APP), as an exemplar of a veterinary pathogen, to identify research gaps relevant to disease prevention and control.
Basic background to APP
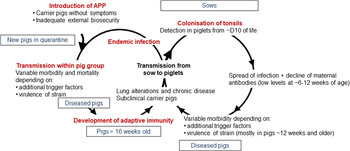
APP is a bacterium that causes pleuropneumonia, a disease of pig lungs that causes substantial morbidity and mortality in the worldwide porcine industry (Bossé et al., Reference Bossé, Janson, Sheehan, Beddek, Rycroft, Kroll and Langford2002; Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019; Gale and Valazquez, Reference Gale and Valazquez2020). The dynamics of infection with APP were recently reviewed and are summarized in Fig. 1 (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018). Pigs, both domesticated and wild (Vengust et al., Reference Vengust, Valencak and Bidovec2006), are the only natural host for APP, and the outcome of host–bacterial interaction is subclinical disease (no clinical signs, no lung lesions at slaughter), chronic disease (low mortality, few and/or low specific clinical signs, reduced growth rate and/or lung lesions at slaughter), acute disease (high or intermediate mortality, lung lesions at slaughter), tonsil colonization (which subsequently spreads to the lungs), or bacterial clearance (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). APP is spread by direct contact and/or by aerosol (Tobias et al., Reference Tobias, Bouma, Daemen, Wagenaar, Stegeman and Klinkenberg2013). Control is through a combination of husbandry, antibiotics, and vaccines (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). However, the emergence of antimicrobial resistance is a major concern (Michael et al., Reference Michael, Bossé, Schwarz, Aarestrup, Schwarz, Shen and Cavaco2018), and the most widely used bacterin (whole killed) vaccines only protect against one or, at best, a few of the 19 known serovars (Bossé et al., Reference Bossé, Li, Sárközi, Fodor, Lacouture, Gottschalk, Casas Amoribieta, Angen, Nedbalcova, Holden, Maskell, Tucker, Wren, Rycroft and Langford2018b; Stringer et al., Reference Stringer, Bossé, Lacouture, Gottschalk, Fodor, Angen, Velazquez, Penny, Lei, Langford and Li2021), and do not prevent colonization (Loera-Muro and Angulo, Reference Loera-Muro and Angulo2018). There is thus considered an urgent need for substantially improved and new prevention and therapeutic strategies.
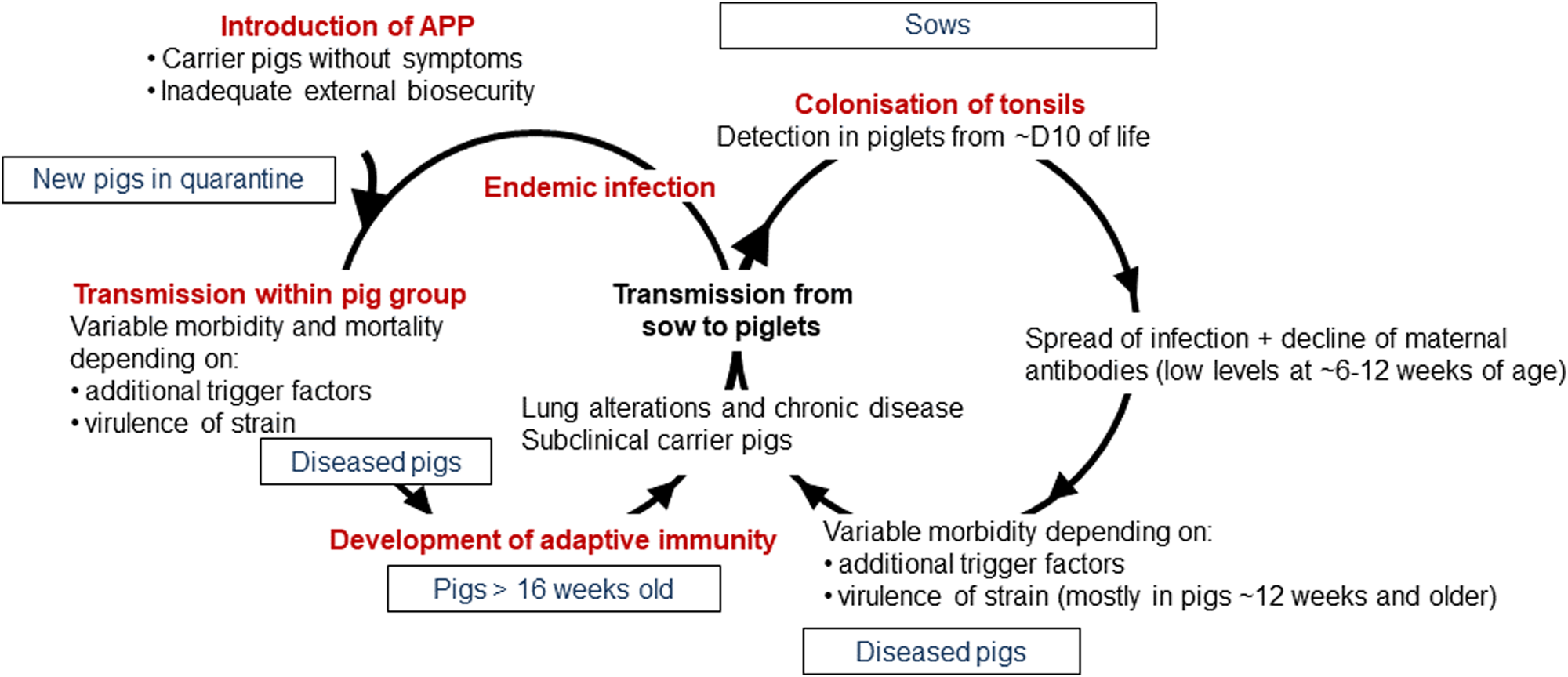
Fig. 1. Dynamics of infection with APP. Adapted with permission from Sassu et al. (Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018). See text for further details.
Here, we show how application of the MISTEACHING framework to the veterinary pathogen APP can be used to identify gaps in our current knowledge, and to formulate hypotheses whose study will enhance prospects for disease control. It should be noted, as pointed out in the original MISTEACHING paper (Casadevall and Pirofski, Reference Casadevall and Pirofski2018), that there is some overlap between the categories, and examples will be mentioned where appropriate. In addition, we propose, and present our reasoning for, an extra category – Strain – to be added to the MISTEACHING framework, so that it becomes MISTEACHINGS.
The MISTEACHINGS framework applied to APP
Microbiome
The two primary niches that APP occupies are the tonsils (Chiers et al., Reference Chiers, Haesebrouck, van Overbeke, Charlier and Ducatelle1999) and the lung (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019), although recent data suggest that multi-organ spread of the bacterium, significantly correlated with spleen colonization, may occur during acute infection following lung colonization (Hoeltig et al., Reference Hoeltig, Rohde, Frase, Nietfeld, Waldmann, Valentin-Weigand and Meens2018). Studies of the tonsil microbiome of pigs that had no history of respiratory diseases and were considered free of APP, showed a post-birth early litter-related microbiome with high similarity to that of the sow teat skin and vagina. However, the tonsil microbiomes of individual litters converged over the following 3 weeks, and dramatically diverged at 4 weeks, with the stresses of a change in diet (weaning), change in room, and addition of in-feed antibiotic (Pena Cortes et al., Reference Pena Cortes, LeVeque, Funk, Marsh and Mulks2018a). Notably, members of the family Pasteurellaceae, of which APP is a member, were present throughout the period, as they were in a longer study of 19 weeks (Pena Cortes et al., Reference Pena Cortes, LeVeque, Funk, Marsh and Mulks2018b). The results were consistent with previous tonsil microbiome studies on 18–20 week healthy pigs (Lowe et al., Reference Lowe, Marsh, Isaacs-Cosgrove, Kirkwood, Kiupel and Mulks2011, Reference Lowe, Marsh, Isaacs-Cosgrove, Kirkwood, Kiupel and Mulks2012), and earlier studies reviewed by Kernaghan et al. (Reference Kernaghan, Bujold and MacInnes2012). APP can transmit from the sow to piglets as early as 10 days (Vigre et al., Reference Vigre, Angen, Barfod, Lavritsen and Sørensen2002). However, the effect of APP colonization on the tonsillar microbiome of healthy pigs is unknown. Not all litter mates will be colonized with APP, although the numbers increase with age (Tobias et al., Reference Tobias, Klinkenberg, Bouma, van den Broek, Daemen, Wagenaar and Stegeman2014b), but whether there are bacterial species/consortia that can prevent tonsil colonization is also unknown, as is the effect of vaccination on the microbiome, and these can be considered as research gaps.
Similarly, there is a little information on the composition of the lung microbiome, either in health or disease. Siqueira et al. carried out a shotgun metagenomic study of lung lavage samples from a herd of pigs kept in field conditions with signs of infection with enzoonotic pneumonia, caused by Mycoplasma hyopneumoniae, or with no signs of infection (Siqueira et al., Reference Siqueira, Pérez-Wohlfeil, Carvalho, Trelles, Schrank, Vasconcelos and Zaha2017). This descriptive study found that Mycoplasmataceae, Flavobacteriaceae, and Pasteurellaceae were the most common families identified in lungs with signs of enzoonotic pneumonia compared to Mycoplasmataceae, Bradyrhizobiaceae, and Flavobacteriaceae in those without signs of infection. A similar 16S rRNA-based microbiome study on lung lavages of slaughter pigs, with or without lung lesions, found microbial diversity was significantly reduced in lungs with severe-lesions compared to those with ‘slight-lesions’ or that were considered healthy (Huang et al., Reference Huang, Zhang, Tong, Chen, Yan, Fang, Guo, Yang, Xiao, Chen, Huang and Ai2019). Mycoplasma and Ureaplasma were enriched in severe-lesion lungs, and 62 Operational Taxonomic Units (OTUs) were negatively associated with the presence of lung lesions, 49 of which were relatively low abundance (<0.05%). A similar 16S rRNA-based study of the lungs of healthy and diseased Duroc × Landrace × Yorkshire crossbreed pigs found a higher relative abundance of Lactococcus, Enterococcus, Staphylococcus, and Lactobacillus in healthy pig lungs and an enhanced richness of Streptococcus, Haemophilus, Pasteurella, and Bordetella genera in diseased lungs, although no diagnostics as to the primary or secondary infections of diseased pigs was reported (Li et al., Reference Li, Wang, Di, Pan, Gao, Xiao, Li, Wei, Liu, Qiu and Ma2020). Another 16S rRNA-based study comparing the diversity of bacteria in the alveolar lavage fluid obtained from healthy and diseased lungs of 4-week-old Kele pigs has also been reported (Zhang et al., Reference Zhang, Shi, Wang, Zhang, Zhao, Du and Zhang2020). Taken collectively, the results suggest that infection of pig lungs with bacterial pathogens can dramatically alter the microbiome, and that colonization with many divergent beneficial bacteria may have a role in protection of the lungs against pathogenic microorganisms. Whether APP infection of the lungs will also lead to a lack of bacterial diversity, and whether the presence of bacterial species/consortia can prevent infection is unknown. APP is a primary pathogen of the porcine respiratory disease complex (PRDC), where infection can involve multiple bacteria and/or viruses, so tonsil and lung microbiome-based studies may need to be monitored for polymicrobial infection, although the bacterium can be the primary agent of lung disease (Opriessnig et al., Reference Opriessnig, Gimenez-Lirola and Halbur2011; Saade et al., Reference Saade, Deblanc, Bougon, Marois-Créhan, Fablet, Auray, Belloc, Leblanc-Maridor, Gagnon, Zhu, Gottschalk, Summerfield, Simon, Bertho and Meurens2020), and initial experimental APP mono-infection studies may provide important baseline data.
Also of interest is the interplay between the pig gut and lung microbiota (the gut−lung axis) and vice versa. Infection of specific pathogen free pigs with APP was associated with a change in the Simpson's diversity index of fecal coliforms, indicating lower diversity in the gut (Zoric et al., Reference Zoric, Arvidsson, Melin, Kühn, Lindberg and Wallgren2010). Additionally, the richness score for taxa Ruminococcus_2 found in the guts of litters correlated significantly with the lowest lung lesions scores of animals experimentally infected with M. hyopneumoniae (Surendran Nair et al., Reference Surendran Nair, Eucker, Martinson, Neubauer, Victoria, Nicholson and Pieters2019a). The results suggest that M. hyopneumoniae susceptibility is associated with early life gut microbiota. Similarly, the presence of non-pathogenic Escherichia coli and increased fecal microbiome diversity in the pig gut was associated with reduced lung pathology after experimental infection with porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circovirus type 2, both agents of the PRDC (Niederwerder, Reference Niederwerder2017). Taken together, these studies suggest the possibility of targeted alteration of the pig gut microbiome, e.g. through probiotic or defined microbial consortia or purified metabolites (Wypych et al., Reference Wypych, Wickramasinghe and Marsland2019), to decrease susceptibility to lung disease caused by infectious agents, and future studies may also include inter-kingdom cross talk between bacteria, fungi and viruses which has been recognized as a component of healthy/diseased lung health (Enaud et al., Reference Enaud, Prevel, Ciarlo, Beaufils, Wieërs, Guery and Delhaes2020). Again, there is no information on whether the gut microbiome is associated with susceptibility to lung disease caused by APP, and the above discussion indicates there is a major gap in knowledge concerning APP and the pig tonsil/lung/gut microbiome.
Immunity
The immune status of an animal will clearly influence susceptibility to APP disease, and overlaps with many other categories in the MISTEACHING(S) framework e.g. Microbiome, Age, Environment, History, Nutrition, Genetics, and Strain. Historically, the main component considered in surviving an encounter with a pathogen – and therefore a component of infectious disease susceptibility – is disease resistance, i.e. the ability of the host to kill a pathogen (Ayres and Schneider, Reference Ayres and Schneider2012). Immune-driven disease resistance mechanisms (including the innate, humoral, and cell-mediated responses) involved in elimination of APP have recently been reviewed in detail (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018) and will not be considered here. Examples demonstrating the importance of immunity, or lack of, to the outcome of APP-host interaction include: the substantial morbidity and mortality that arises when naïve animals, with/without preceding infection and/or co-infection with viruses, come into contact with asymptomatic carriers (Opriessnig et al., Reference Opriessnig, Gimenez-Lirola and Halbur2011), and that vaccination with bacterins can reduce severity of lung disease (Loera-Muro and Angulo, Reference Loera-Muro and Angulo2018). In general, such disease resistance mechanisms are poorly understood, and this can be considered a major research gap that needs addressing to formulate new prevention and/or treatment strategies.
In addition to disease resistance, it is now recognized that disease tolerance, i.e. the ability to maintain host fitness without affecting pathogen burden, is also an important determinant of host susceptibility to infectious disease (Soares et al., Reference Soares, Teixeira and Moita2017). While the mechanisms that are involved in disease tolerance are not fully understood, they include tissue damage control mechanisms based on evolutionarily conserved stress and damage responses (Soares et al., Reference Soares, Teixeira and Moita2017), and host metabolism and immune crosstalk (McCarville and Ayres, Reference McCarville and Ayres2018). There can be considerable tissue damage during acute APP infection. Apx toxins and the induced inflammatory response can result in substantial lung damage, which can lead to chronic disease where characteristic abscess-like nodular lesions surrounded by connective tissue are a characteristic gross pathological finding (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). Thus, tissue damage control mechanisms are apparent in the response to APP infection. Also associated with chronic disease is the persistence of APP in tonsillar crypts (Müllebner et al., Reference Müllebner, Sassu, Ladinig, Frombling, Miller, Ehling-Schulz, Hennig-Pauka and Duvigneau2018). Such animals maybe asymptomatic but are still capable of transmitting APP to naïve animals (Chiers et al., Reference Chiers, De Waele, Pasmans, Ducatelle and Haesebrouck2010). The host and bacterial mechanisms that allow APP to survive in the tonsillar crypts are poorly understood, and have significant control implications. A recent study analyzed cytokine responses in German Landrace pigs that had been infected intranasally with a serovar 2 strain of APP (Müllebner et al., Reference Müllebner, Sassu, Ladinig, Frombling, Miller, Ehling-Schulz, Hennig-Pauka and Duvigneau2018), and identified that acute disease was associated with increased expression of the pro-inflammatory interleukin-17A (IL-17A) in the lung, levels of which had previously been shown to correlate with the presence of lung lesions in chronically infected pigs (Sassu et al., Reference Sassu, Ladinig, Talker, Stadler, Knecht, Stein, Frömbling, Richter, Spergser, Ehling-Schulz, Graage, Hennig-Pauka and Gerner2017). It is known that Th17 immunopathology is largely driven by products of neutrophil activation, such as reactive oxygen species and elastase (Soares et al., Reference Soares, Teixeira and Moita2017), and has parallels with work suggesting that Th17 responses are important in protection against human respiratory pathogens, e.g. Haemophilus influenzae (Noda et al., Reference Noda, Kodama, Umemoto, Nomi, Hirano and Suzuki2011), and Streptococcus pneumoniae (Ramos-Sevillano et al., Reference Ramos-Sevillano, Ercoli and Brown2019). In contrast, in APP-chronically infected tonsils there was increased expression of the anti-inflammatory cytokine IL-10. Indeed, in both lung and tonsil, there was a marked reciprocal correlation between IL-17A and IL-10 concentrations. IL-10 is considered a master regulator of immunity to infection, inhibiting macrophages, Th1 and natural killer cells, which can result in impedance of pathogen clearance at the expense of preventing excessive tissue damage (Couper et al., Reference Couper, Blount and Riley2008). APP-infected pigs pre-treated with adenovirus-5 expressing human IL-10 had a significant reduction in lung damage compared to controls (Morrison et al., Reference Morrison, Foss and Murtaugh2000). Müllebner et al. hypothesized that ‘APP adapts its metabolism to trigger IL-10 production and consequently facilitates chronic APP persistence inside porcine tonsillar tissue’ (Müllebner et al., Reference Müllebner, Sassu, Ladinig, Frombling, Miller, Ehling-Schulz, Hennig-Pauka and Duvigneau2018). From a bacterial perspective, the hypothesis is supported by differences found in Fourier transform infrared spectroscopy of serovar 2 isolates from lung and tonsillar tissue (Aper et al., Reference Aper, Frömbling, Bağcıoğlu, Ehling-Schulz and Hennig-Pauka2020). That respiratory pathogens can induce IL-10 and control of tissue damage, an example of diseases tolerance, is exemplified by Staphylococcus aureus (Chau et al., Reference Chau, McCully, Brintnell, An, Kasper, Vines, Kubes, Haeryfar, McCormick, Cairns, Heinrichs and Madrenas2009). Further work is required to identify any APP factors involved in IL-10 induction and the host interactive biology, and such knowledge can potentially be used in formulating strategies to eliminate APP tonsillar carriage.
Also pertinent to disease tolerance is APP outbreaks associated with stress-inducing trigger factors in already colonized but asymptomatic animals (see Environment). Homeostasis (i.e. the ability of an entire organism, organ, or individual cell to maintain key regulated variables within an acceptable range) is a fundamental property of biological systems (Chovatiya and Medzhitov, Reference Chovatiya and Medzhitov2014). Such parameters include oxygen, pH, glucose, and ATP. When these change beyond a certain threshold, host cells are alerted by stress sensors, such as pathogen-associated molecular pattern molecules (PAMPs) and damage-associated molecular patterns, and induce signal transduction pathways that can restore homeostasis. Receptors sensing general environmental stress (and infection) typically involve signaling through nuclear factor (NF)-κB, mitogen-activated protein kinases (MAPKs), and cJun NH2-terminal kinase (JNK) (Chovatiya and Medzhitov, Reference Chovatiya and Medzhitov2014). In porcine alveolar macrophages, ApxI is known to induce apoptosis via MAPKs p38 and JNK (Wu et al., Reference Wu, Chen, Chen, Liao, Lin, Chien, Lee and Hsuan2011), and IL-1β, IL-8, and TNF-α production via NF-κB in a JNK-dependent manner (Hsu et al., Reference Hsu, Li, Chang, Chen, Liao, Chen, Wang, Lin and Hsuan2016). Metabolic adaptation to stress in host cells confers tissue damage control (Soares et al., Reference Soares, Teixeira and Moita2017), in part by releasing metabolites, e.g. ATP, that influence innate immunity (Naquet et al., Reference Naquet, Giessner and Galland2016) contributing to disease tolerance control (Soares et al., Reference Soares, Teixeira and Moita2017). There is little information on tonsillar cell responses induced by stress triggers, but in transcriptomic studies of acute APP-infected lungs it was noteworthy that differentially expressed genes in necrotic areas were largely those involved in regulation of homeostasis (Mortensen et al., Reference Mortensen, Skovgaard, Hedegaard, Bendixen and Heegaard2011). Understanding disease tolerance mechanisms in the context of APP colonization of tonsils has the potential for the formulation of therapeutic strategies that target stress and damage responses. Proof-of-principle that the general concept of targeting disease tolerance can work is shown by the prevention of tissue damage and lethality from sepsis following administration of the heme-sequestering protein, hemopexin, to mice (Larsen et al., Reference Larsen, Gozzelino, Jeney, Tokaji, Bozza, Japiassu, Bonaparte, Cavalcante, Chora, Ferreira, Marguti, Cardoso, Sepulveda, Smith and Soares2010; Soares et al., Reference Soares, Teixeira and Moita2017). Elimination of APP from the tonsils is notoriously difficult (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018), and currently complete restocking with APP-free animals is considered the only certain way of preventing the spread of infection. We believe that approaches to eliminate APP tonsillar colonization by immunomodulators targeting disease resistance (with or without antibiotics) is a worthy area of research. In addition, symbiotic microbiome-based approaches to reduce or prevent pathogen load (see Microbiome) could be considered as targeting disease tolerance, if they can be shown experimentally to induce immunoregulatory mechanisms and/or involve stress and damage responses (Soares et al., Reference Soares, Teixeira and Moita2017).
Tissue damage control is also relevant to co-infections (Soares et al., Reference Soares, Teixeira and Moita2017) and is considered a disease tolerance mechanism. Co-infection of an APP serovar 2 and an H1N1 influenza strain resulted in more severe symptoms and lung lesions and enhanced viral replication in the lung and nasal shedding, compared to pigs infected with single agents (Pomorska-Mól et al., Reference Pomorska-Mól, Dors, Kwit, Kowalczyk, Stasiak and Pejsak2017). The mechanisms by which particular viruses or bacteria compromise disease tolerance to secondary infections is likely to be multifactorial, with a degree of specificity for the combinations involved, but have been associated with T-regulatory (Treg) cell and amphiregulin dysregulation (Soares et al., Reference Soares, Teixeira and Moita2017; McCarville and Ayres, Reference McCarville and Ayres2018). Again, disease resistance and tolerance mechanisms preventing/limiting APP infection in the context of co-infections is a research gap with a need for more experimentally controlled studies, as undertaken by Pomorska-Mól (Pomorska-Mól et al., Reference Pomorska-Mól, Dors, Kwit, Kowalczyk, Stasiak and Pejsak2017).
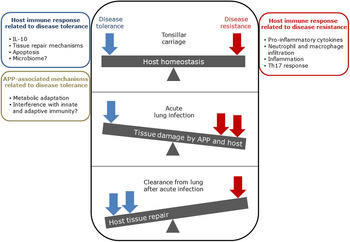
The balance between disease resistance and tolerance will determine APP load and host damage (Fig. 2). However, this can be difficult to experimentally determine as: (1) the same cell types, e.g. macrophages, are involved in both disease resistance and tolerance; and (2) it often requires disease resistance to be stably maintained, e.g. through the use of antibiotics (Soares et al., Reference Soares, Teixeira and Moita2017). One approach is to plot disease (health) parameters versus pathogen load over time, and the fitness curves can potentially be used to estimate variations in disease tolerance (Schneider, Reference Schneider2011). While APP load is relatively easy to quantify, choice of an informative health parameter to measure is considerably more difficult (Schneider, Reference Schneider2011). Nevertheless, such disease curves have the potential to identify parts of the APP disease process that are worthy of exploration. An alternative way of thinking, that reflects the overlap between disease tolerance and resistance, is to consider inflammation as the extreme end of a spectrum that ranges from hemostasis to the stress response to the inflammatory response (Chovatiya and Medzhitov, Reference Chovatiya and Medzhitov2014). In this model, inflammation can be triggered either by: (1) extreme deviation of regulated variables (e.g. temperature) from normal values, and is classed as a stress response; or (2) a challenge (e.g. infections, toxins, or tissue damage) that can cause deviation of regulated variables, but is not itself a regulated variable, and is classed as a defense response. Thus, inflammation has both stress and defense response components.
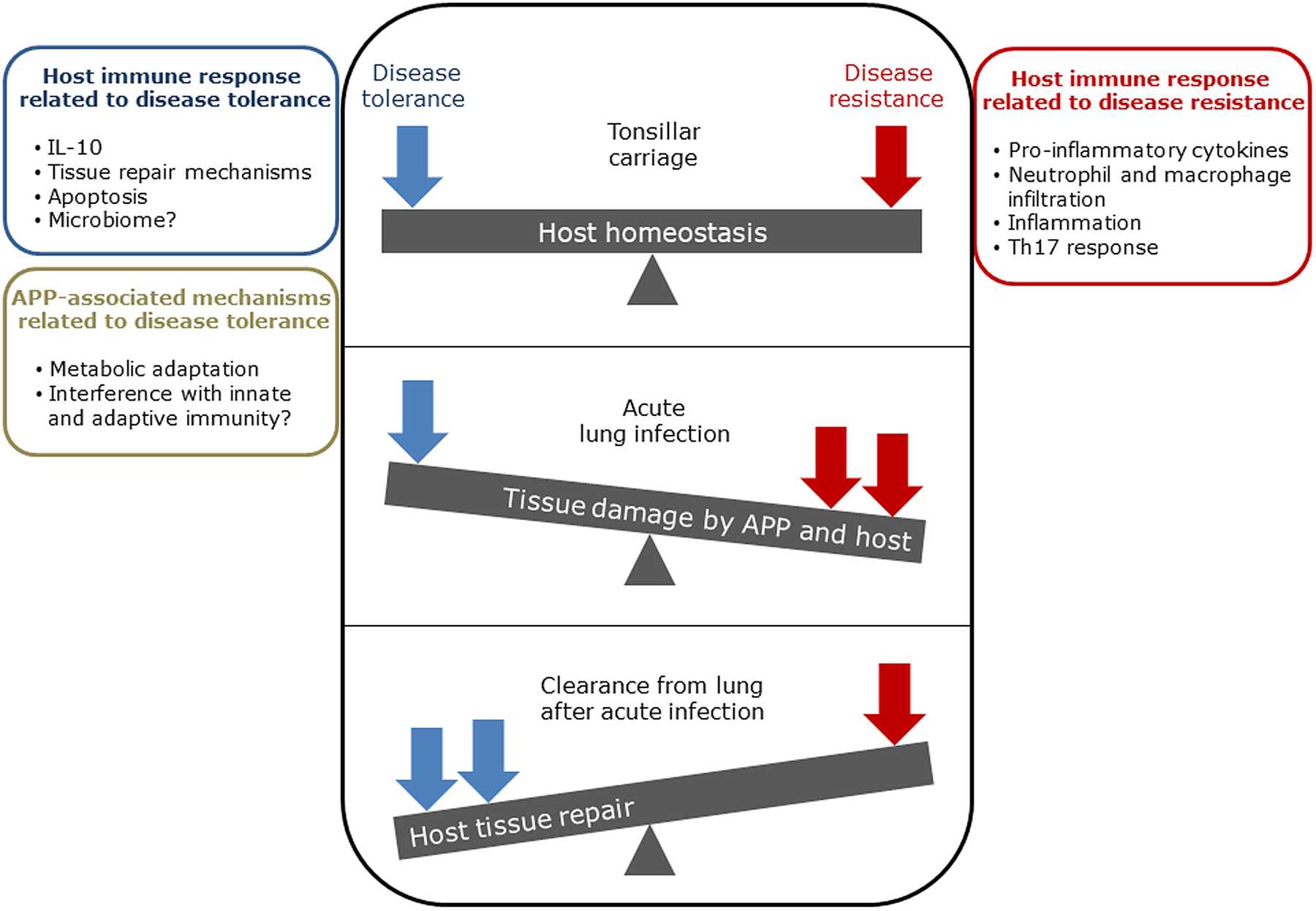
Fig. 2. Schematic representation of the balance between disease tolerance (blue, LHS) and resistance (red, RHS) in relation to APP. Adapted with permission from Shourian and Qureshi (Reference Shourian and Qureshi2019). TOP: During asymptomatic carriage there is a balance between host tolerance and resistance mechanisms, and infection is controlled. CENTRE: During acute disease initiated by stress events to asymptomatic carriers or acquisition of bacteria by naïve hosts, there is considerable damage to the lung caused directly by Apx toxins and also from the host. BOTTOM: Subsequently, APP is cleared from the lung and the host repairs the damage mediated by both the bacterium and host in fighting acute infection. At this stage APP can colonize the tonsils asymptomatically (TOP), and the cycle continues. See text for further details.
In summary, the basic immunology underlying APP-host interactive biology, and the contribution of disease tolerance to host control of APP, especially during chronic infection, can be considered as major research gaps.
Sex
That susceptibility to disease caused by a pathogen can be different between sexes (arising from anatomical, immune function, and genetic differences) is well established (Klein and Flanagan, Reference Klein and Flanagan2016; vom Steeg and Klein, Reference vom Steeg and Klein2016). However, susceptibility to APP appears to be similar in both sexes in a variety of breeds (Straw et al., Reference Straw, Neubauer and Leman1983), and postweaning mortality in commercial swine production was not sex-dependent (Gebhardt et al., Reference Gebhardt, Tokach, Dritz, DeRouchey, Woodworth, Goodband and Henry2020). Furthermore, genetic studies of susceptibility and resistance of pigs to APP, found no differences between sexes (Reiner et al., Reference Reiner, Bertsch, Hoeltig, Selke, Willems, Gerlach, Tuemmler, Probst, Herwig, Drungowski and Waldmann2014a). However, although available data suggests that sex does not appear to make any apparent significant contribution to susceptibility/resistance to APP-induced disease, the mechanisms involved may be different between the sexes, particularly when stressors are involved. For example, neurocrine, immunologic, and behavioral responses of maternal- and littermate-deprived German Landrace piglets challenged with lipopolysaccharide (LPS), as a surrogate for infection, found sex differences between the groups (Brückmann et al., Reference Brückmann, Tuchscherer, Tuchscherer, Gimsa and Kanitz2020). Plasma TNF-α and IL-6 concentrations of LPS-challenged control piglets were significantly higher in males than in females. Females also showed significantly higher expression of amygdala anti-inflammatory IL-10 levels when compared to all comparable male groups, possibly explaining the lower peripheral TNF-α and IL-6 concentrations in females. It was concluded that early life maternal-deprivation alters neuroendocrine and immune responses to acute endotoxaemia in a sex-specific manner.
In addition, from a bacterial perspective, it is known that bacteria can respond to and metabolize sex hormones (reviewed in vom Steeg and Klein, Reference Vom Steeg and Klein2017). For example, many gut bacteria produce hydroxysteroid hydrogenases, regulating the balance between active and inactive steroids (García-Gómez et al., Reference García-Gómez, González-Pedrajo and Camacho-Arroyo2013). While there has not been, to our knowledge, any description of APP metabolizing sex hormones, the closely related bacterium Aggregatibacter actinomycetemcomitans, which occupies the oral cavity of humans, can reduce testosterone to 5α-dihydrotestosterone (Soory, Reference Soory1995). Microbiome composition can affect the response to vaccines (Ferreira et al., Reference Ferreira, Antunes and Finlay2010), and is sex-dependent (Klein et al., Reference Klein, Marriott and Fish2015). This link has the potential for exploitation to maximize vaccine efficacy, which, for APP, may inform design of vaccines to prevent sow colonization.
Temperature
Fever (40.5–41 °C) is a clinical sign in acute disease cases due to APP (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). Induction of fever, in response to infection and inflammatory disease, is a trait of warm- and cold-blooded vertebrates, conserved for over 600 million years (reviewed in Evans et al., Reference Evans, Repasky and Fisher2015). The evidence suggests that, in general, a mild fever is considered of potential benefit but excessive fever maladaptive. The induction and maintenance of fever is known to involve a complex interaction of the innate immune system with neuronal circuits driven by the IL-6-COX2-PGE2 axis (Bernheim et al., Reference Bernheim, Block and Atkins1979; Evans et al., Reference Evans, Repasky and Fisher2015). The process is initiated after binding of PAMPs, e.g. LPS, to pathogen recognition receptors (PRRs), e.g. Toll-like receptor 4 (TLR4). In the case of APP, LPS-induced fever in rabbits (Maudsley et al., Reference Maudsley, Kadis and Mayberry1986), and endobronchial inoculation with ApxI and III, and to a lesser extent ApxII, induced fever in pigs (Kamp et al., Reference Kamp, Stockhofe-Zurwieden, van Leengoed and Smits1997). Correlational antipyretic and hyperthermic/hypothermic studies suggest that fever is an adaptive response enhancing specific and non-specific immunity (Bernheim et al., Reference Bernheim, Block and Atkins1979). For example, in experimental infection studies of New Zealand rabbits with Pasteurella multocida (like APP, a member of the Pasteurellaceae) there was a statistically significant correlation between fever magnitude and survival. An increase in fever of up to 2.25 °C was associated with an improved survival rate, but further increases with reduced survival rate. There was no difference in growth rate in vitro of P. multocida at normal (39 °C) and febrile (42 °C) temperatures, suggesting no direct inhibition of bacterial growth at increased temperature in rabbits, and the results were attributed to ‘an enhancement of some aspect of the rabbits' immunological defences’ (Kluger and Vaughn, Reference Kluger and Vaughn1978). APP can grow at 42 °C for at least 8 h in vitro in rich medium supporting growth (Xie et al., Reference Xie, Zhang, Li, Zhou, Liu and Wang2013), thus it is likely (by analogy to P. multocida) that fever results in immune enhancement rather than direct killing during acute APP infection. Practically, detection of the rise in temperature associated with APP infection by thermal infrared imaging techniques has been evaluated as a diagnostic (Menzel et al., Reference Menzel, Beyerbach, Siewert, Gundlach, Hoeltig, Graage, Seifert, Waldmann, Verspohl and Hennig-Pauka2014; Jorquera-Chavez et al., Reference Jorquera-Chavez, Fuentes, Dunshea, Warner, Poblete, Morrison and Jongman2020). As pointed out by Casadevall (Reference Casadevall2016), there are many studies that have correlated fever with survival, but none that unambiguously show causation. In part, this is due to the difficulty in determining how change in temperature affects both the immune system and the pathogen. Variation in temperature can also act as a stress trigger, increasing susceptibility to disease, and therefore overlaps with Environment and Immunity (see those sections for further details).
In the original MISTEACHING paper, temperature was considered from the host, but not bacterial, perspective. For many pathogens host body temperature is an environmental signal to induce the expression of virulence genes necessary for survival (Shapiro and Cowen, Reference Shapiro and Cowen2012; Lam et al., Reference Lam, Wheeler and Tang2014). Typically, APP is grown in culture at 37 °C, but the bacterium can survive in the environment at room temperature for 3–4 days in the presence of mucin and salt, and on hydrophobic surfaces either under dry or saturated humidity conditions (Assavacheep and Rycroft, Reference Assavacheep and Rycroft2013). APP was cultured from farm drinking water in Mexico and, in data not shown, it was reported that the bacterium could survive in water for at least 3 weeks at 20 °C in the laboratory (Loera-Muro et al., Reference Loera-Muro, Jacques, Tremblay, Avelar-Gonzalez, Loera Muro, Ramirez-Lopez, Medina-Figueroa, Gonzalez-Reynaga and Guerrero-Barrera2013). In addition, the authors detected the presence of APP in biofilm-like structures in the environment. It is not known whether APP found in water or environmental biofilms are as transmissible as those in aerosols or carried by fomites, and their genotype/phenotype (especially in respect to virulence factors) has not been evaluated. Such factors may influence indirect transmission and susceptibility to disease.
Environment
Environmental factors are well established as important determinants of susceptibility of pigs to respiratory disease – see the comprehensive review of Stark (Reference Stark2000). Risks factors associated with respiratory disease of pigs include purchase policy, herd type and size, husbandry system, stocking density, ventilation, air parameters (e.g. temperature), aerosol concentration, risks of contact and airborne infection, and hygiene (Stark, Reference Stark2000; Neumann and Hall, Reference Neumann, Hall, Zimmerman, Karriker, Ramirez, Schwartz, Stevenson and Zhang2019). Appropriate precautions and biosecurity measures should be put in place to reduce the risk of respiratory disease outbreaks. In particular, good herd management practices are necessary to reduce acute stress, e.g., from mixing, moving or weaning – so called ‘triggers’ of stress. Observational studies have indicated a link between changing environment and outbreaks of APP disease, suggesting an external trigger that results in an altered pig−APP interaction (Klinkenberg et al., Reference Klinkenberg, Tobias, Bouma, van Leengoed and Stegeman2014). Such a change could arise due to effects on pig homeostasis and/or the immune system (see Immunity), and/or the phenotype of the bacterium. Knowing whether clinical cases result from an external trigger in colonized pigs (trigger mechanism), or if an initial case precipitates an outbreak (transmission mechanism), is important in formulating optimal control strategies (Klinkenberg et al., Reference Klinkenberg, Tobias, Bouma, van Leengoed and Stegeman2014). A simulation using parameters derived from the literature suggested that APP outbreaks primarily arise from the trigger mechanism, with clear control implications, but that the relationship between subclinical lung lesions, antibodies, protection and disease warranted further study (Klinkenberg et al., Reference Klinkenberg, Tobias, Bouma, van Leengoed and Stegeman2014). Although one study (Maes et al., Reference Maes, Chiers, Haesebrouck, Laevens, Verdonck and de Kruif2001) indicated that APP serovar may play a role in how risk factors (such as origins of purchased pigs and biosecurity) affect outcome of infection, it is noteworthy that there are few published studies directly addressing the effect of housing system on prevalence of APP disease. Temperature was considered as a risk factor for respiratory diseases in pigs bred outdoors (Beskow et al., Reference Beskow, Norqvist and Wallgren1998) and in units with natural, compared to mechanical, ventilation (Chantziaras et al., Reference Chantziaras, De Meyer, Vrielinck, Van Limbergen, Pineiro, Dewulf, Kyriazakis and Maes2020). However, in general, respiratory disease is less prevalent in pigs reared outdoors compared to indoors (Delsart et al., Reference Delsart, Pol, Dufour, Rose and Fablet2020). Social and environmental enrichment had an impact on ‘disease susceptibility’ of pigs infected initially with PRRSV and 8 days later with a serovar 2 APP isolate (van Dixhoorn et al., Reference van Dixhoorn, Reimert, Middelkoop, Bolhuis, Wisselink, Groot Koerkamp, Kemp and Stockhofe-Zurwieden2016). Pigs in enriched conditions showed less stress behavior, and reduced disease susceptibility. In relation to APP, pigs raised in pens under enriched conditions had fewer lung lesions (7.1% of animals) compared to those raised in barren conditions (57% of animals). The authors suggested that the results supported the effect of housing on the hypothalamic−pituitary−adrenal (HPA) axis. Stress hormones epinephrine and norepinephrine are known to alter expression of APP genes, including those encoding virulence factors (Li et al., Reference Li, Xu, Zhou, Sun, Liu, Chen and Zhou2012), particularly those involved in regulation of growth, iron-acquisition and metabolism (Li et al., Reference Li, Chen, Bei, Su, Huang, Zhang, Chen and Zhou2015), thereby linking with nutritional immunity (see Nutrition section). In summary, the evidence suggests that there is a clear link between environment and respiratory diseases in pigs, however this is an area where more controlled experiments would facilitate more robust correlations with disease caused by APP.
Age
Transmission from infected sows to piglets is known to occur from 10 days onwards, although not all litter mates, or all litters, are infected at the same time (Vigre et al., Reference Vigre, Angen, Barfod, Lavritsen and Sørensen2002). Suckling pigs rarely develop disease, especially if maternally derived antibodies provide protection. Maternal antibodies wane and are generally below detectable limits by 12 weeks of age (dependent on the test used), after which there is higher risk of disease (reviewed in Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018). However, in naïve pigs, it has been reported that susceptibility to aerosol infection with APP was greater at 10 compared to 12 weeks of age, although no explanation was given for the results (Sebunya et al., Reference Sebunya, Saunders and Osborne1983). Thus, in pig production systems worldwide, APP infection is predominantly a disease of pigs of 10 or more weeks of age. There are many unanswered questions relating to the immune mechanisms in sows versus those in piglets, especially relating to the nature and targets of maternal antibodies conferring protection against colonization. Understanding such mechanisms has clear implications for formulating sow and/or piglet-based strategies to prevent APP disease.
Chance
Many of the categories in the MISTEACHING framework of host susceptibility can be considered as stochastic, e.g. History, Inoculum, and Genetics (Casadevall and Pirofski, Reference Casadevall and Pirofski2018). It could be argued that the element of chance of APP infection can be minimized in well run pig units, as the environment (e.g. temperature, ventilation, biosecurity, stocking density, etc.) can be tightly controlled. Transmission from pig to pig is by direct oral or nasal contact, or via aerosols over 1–2 m (Nicolet et al., Reference Nicolet, Konig and School1969; Kristensen et al., Reference Kristensen, Angen, Andreasen, Takai, Nielsen and Jorsal2004), with direct contact being 10× more efficient than indirect transmission (Tobias et al., Reference Tobias, Bouma, van den Broek, van Nes, Daemen, Wagenaar, Stegeman and Klinkenberg2014a). There are, however, reported cases of indirect aerosol transmission between independent pig units (different organizations, no known contact between units) over distances of 500 m (Desrosiers and Moore, Reference Desrosiers and Moore1998; Larsen, Reference Larsen1998). Prior to one outbreak (case 2), weather reports indicated that the dominant winds were from the APP-infected farm to the one with no history of infection with the bacterium (Desrosiers and Moore, Reference Desrosiers and Moore1998). The strains involved were of the same serovar and antimicrobial sensitivity pattern. While identical serovar and antimicrobial sensitivity do not definitely prove the strains were the same, the data suggests the possibility of indirect transmission between unrelated units which, given the circumstances, could be considered as a chance event. With the advent of next generation sequencing technology and the application of single-nucleotide polymorphism (SNP) analyses, it is now be possible to determine whether APP is transmissible over distances such as 500 m, as in case 2.
SNP analysis also facilitates detection of random genetic mutations, which – by their very nature – are chance occurrences, though environmental pressures selecting for persistence of the mutations can be controlled. In the host, random mutations can affect determinants of susceptibility and severity of infection, which through genetic mapping and selective breeding can be used to improve resistance to specific diseases (see Immunity and Genetics). In bacteria, random mutations typically occur at frequencies between 10−7 and 10−10, but may be higher in the absence of functional mismatch repair (Lynch et al., Reference Lynch, Ackerman, Gout, Long, Sung, Thomas and Foster2016; Chevallereau et al., Reference Chevallereau, Meaden, van Houte, Westra and Rollie2019). In addition to point mutations, spontaneous deletions or rearrangements in bacteria may be mediated by the presence of repeat elements in the chromosome that facilitate recombination, and horizontal gene transfer can lead to acquisition of new genes (Darmon and Leach, Reference Darmon and Leach2014). APP is a naturally transformable bacterium and therefore horizontal gene transfer can occur (Bossé et al., Reference Bossé, Sinha, Schippers, Kroll, Redfield and Langford2009). That chance events can alter phenotype is shown by inactivation of the expression of APP ApxIV (Tegetmeyer et al., Reference Tegetmeyer, Jones, Langford and Baltes2008), as well as capsule and O-polysaccharide (To et al., Reference To, Teshima, Kon, Yasuda, Akaike, Shibuya, Nagai and Sasakawa2020) of APP isolates, by insertion of ISApl1 into their cognate genes. Regardless of the mechanism, mutations which negatively affect fitness tend to be rapidly lost, whereas those increasing fitness become fixed in populations, giving rise to different lineages which may vary in virulence (see Strain, below). It should be noted that improved bacterial fitness does not necessarily equate to increased pathogenicity, as loss of virulence factors may be associated with increased persistence, due to reduced provocation of host immune responses. Part of the lung pathology in acute APP infection is due to release of toxic oxygen metabolites and inflammatory mediators by pulmonary macrophages activated by Apx toxin and LPS stimulation (Chen et al., Reference Chen, Chien, Chang, Chen, Wu, Huang, Lee and Hsuan2011; Li et al., Reference Li, Fang, Zuo, Yin, He, Yang, Deng, Shen, Ma, Yu, Wang and Ren2018), and deletion or insertional inactivation of these and other virulence genes by mobile genetic elements, such as ISApl1 (see above), are documented. Furthermore, in addition to loss or insertional inactivation of genes, acquisition of antimicrobial and/or other resistance genes by horizontal transfer (Michael et al., Reference Michael, Bossé, Schwarz, Aarestrup, Schwarz, Shen and Cavaco2018) may also enhance persistence without increasing pathogenicity. In summary, chance events may influence susceptibility to disease, but these are typically difficult to determine.
History
There is clearly overlap between History and other categories in the MISTEACHING framework, particularly Microbiome, Immunity, and Environment. Heterologous immunity, i.e. infection with one microbe affecting the outcome of infection with a related one through changes in host immunological state, is a feature of APP. As a primary member of the PRDC, APP can itself alter the host immunological state, such that the pig is more or less susceptible to infection by other bacteria and viruses, and vice versa (Opriessnig et al., Reference Opriessnig, Gimenez-Lirola and Halbur2011). This phenomenon can be exploited for disease control. For example, a serovar 1 triple mutant of APP that expressed non-toxic but immunogenic ApxI, ApxII and ApxIV, induced significant protection in pigs against a serovar 5 isolate of Glaesserella (Haemophilus) parasuis (Fu et al., Reference Fu, Ou, Zhang, Xu, Liu, Liu, Yuan, Chen and Bei2013). History is considered an important facet of control of APP. Many pig production units will choose to tolerate the presence of APP, providing that that there are not serious outbreaks of acute disease. As such, APP is endemic in many countries, e.g. 98.2% of herds in Ireland were recently reported serologically positive for APP (Rodrigues da Costa et al., Reference Rodrigues da Costa, Fitzgerald, Manzanilla, O'Shea, Moriarty, McElroy and Leonard2020).
The greatest risk of acute disease is from the introduction of asymptomatic carriers into a herd, exacerbated by the difficulty in identifying such animals (Gottschalk, Reference Gottschalk2015). Once in a herd, APP is difficult to eradicate. Options include: off-site segregated medicated early weaning supported by a program of vaccination; medication; culling and repopulation with disease-free gilts; on-site medicated early weaning; and ‘test and removal’ of seropositive sows under medication (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). Destocking and repopulation from herds which are certified free, or have no history, of APP infection is considered optimal. However, this is expensive and may lead to the loss of blood lines (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019), and once APP-free, strict biosecurity is required to maintain the status. History also includes prior vaccination. The most widely used bacterin (whole cell killed) vaccines reduce the extent of lung lesions, do not prevent colonization, and only protect against homologous or closely related serovars. ApxI-III-based vaccines are aimed at neutralizing the effects the of the various toxin combinations produced by different serovars, and there is considerable research in the area of live-attenuated vaccines, because of the potential to cross-protect against many serovars (reviewed in Loera-Muro and Angulo, Reference Loera-Muro and Angulo2018). Appropriate prior vaccination will reduce the susceptibility of pigs to APP-caused disease.
Inoculum
The effect of APP inoculum size in experimentally infected pigs has been the subject of many studies aimed at understanding host−APP interactive biology, and/or the efficacy testing of vaccines and therapeutics (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018). Both dose and route of administration can influence the outcome of infection. For example, pigs inoculated either intranasally (IN) or endotracheally (ET) with a serovar 1 clinical isolate resulted in infection (Baarsch et al., Reference Baarsch, Foss and Murtaugh2000). However, IN and ET inoculations were respectively associated with unilateral and bilateral gross lesions, and with clinical signs apparent between 6 and 8 h and <2 h. It was concluded that the ET route was superior for experimental infection, as all pigs were infected (unlike the IN route, where only 25% of animals had clinical signs 20 h post inoculation) and resulted in bilateral lesions characteristic of natural infections. With the same APP serovar 2 isolate, 103 colony-forming units (CFU) and 4.9 × 104 CFU of intratracheal and IN doses, respectively, induced clinical signs and lung lesions, but not death (Hennig-Pauka et al., Reference Hennig-Pauka, Baltes, Jacobsen, Stratmann-Selke, Gerlach, Selbitz and Waldmann2008). Endobronchial inoculation with the reference serovar 9 strain CVJ13261, in doses ranging from 8 × 101 to 9 × 107 CFU, identified a unimodal relationship with the extent of clinical symptoms and severity of lesions. A dose of 104 CFU was associated with the highest mortality and severest pneumonic lesions, while no death and less severe lesions were associated with a dose of 106 CFU (van Leengoed and Kamp, Reference van Leengoed and Kamp1989). Indirect transmission via aerosols is an established route of APP infection of pigs, and an aerosol dose relationship study in pigs with serovar 1 strain A79-9 found a correlation between inhaled dose and mortality, with LD50s of 7.0 × 103 and 1.9 × 105 CFU ml−1 in the two separate experiments (Sebunya et al., Reference Sebunya, Saunders and Osborne1983). In general, it was concluded that ET and endobronchial administration enable severe lung infection with a precise bacterial dose, but IN infection tends to lead to milder symptoms and chronic infection, in part due to the loss of the inoculum by coughing and swallowing (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018). In summary, APP infection is influenced by strain, dose and administration route.
Nutrition
That nutrition impacts immunity in pigs is well established (see excellent reviews of Liu et al., Reference Liu, Espinosa, Abelilla, Casas, Lagos, Lee, Kwon, Mathai, Navarro, Jaworski and Stein2018; Pluske et al., Reference Pluske, Kim and Black2018; Bouwens and Savelkoul, Reference Bouwens, Savelkoul, Hendriks, Verstegen and Babinszky2019). However, there has been little research done to predict the optimal diet for immune function in pigs, and such diets may differ from those that are required to avoid deficiencies (Chase and Lunney, Reference Chase, Lunney, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). That diet can affect the response of pigs to APP and other respiratory pathogens of pigs has been described (Turek et al., Reference Turek, Schoenlein, Watkins, Van Alstine, Clark and Knox1996; Becker et al., Reference Becker, van Wikselaar, Mul, Pol, Engel, Wijdenes, van der Peet-Schwering, Wisselink and Stockhofe-Zurwieden2012; Surendran Nair et al., Reference Surendran Nair, Eucker, Martinson, Neubauer, Victoria, Nicholson and Pieters2019a). For example, compared to control pigs fed an un-supplemented diet, those fed a diet supplemented with 5% garlic showed a lower incidence of lung lesions following aerosol-delivered APP serovar 2 (Becker et al., Reference Becker, van Wikselaar, Mul, Pol, Engel, Wijdenes, van der Peet-Schwering, Wisselink and Stockhofe-Zurwieden2012). Experimental infection studies in pigs fed diets with different polyunsaturated fatty acids (PUFAs) found that there was a relationship between the content of (n−3):(n−6) PUFAs in alveolar macrophages and outcome of M. hyopneumoniae infection (Turek et al., Reference Turek, Schoenlein, Watkins, Van Alstine, Clark and Knox1996). High-resolution liquid chromatography–mass spectrometry (LC–MS) analysis also found significantly increased serum α-aminobutyric acid and long-chain fatty acids at 14 and 21 days post M. hyopneumoniae infection (Surendran Nair et al., Reference Surendran Nair, Yao, Chen and Pieters2019b). There is a lack of well-controlled studies regarding the effect of diet on the susceptibility of pigs to APP infection, especially those combining metabolomic analyses such as those by Surendran Nair et al. (Reference Surendran Nair, Yao, Chen and Pieters2019b), which have the potential to rapidly advance our understanding and to improve control measures. Provision of feed for pigs is a major contributor to land and water use and greenhouse gas emissions, and sustainable environmentally friendly produced feed sources, such as insects, are under increasing investigation (DiGiacomo and Leury, Reference DiGiacomo and Leury2019), and these have been associated with higher IgG blood levels compared to a conventional diet (Ko et al., Reference Ko HS, Kim and Kim2020). This is an area which can be considered a major knowledge gap, and it is likely that future research will evaluate environmentally sustainable food approaches on the incidence of APP and other respiratory diseases in pigs.
Although not considered in the original MISTEACHING framework, nutritional immunity can be considered under this category, in addition to overlapping with Immunity. The phrase nutritional immunity was first introduced by Weinberg (Reference Weinberg1975), and refers to a host sequestering trace minerals, such as iron and zinc, to limit disease progression and severity after infection (reviewed in Hood and Skaar, Reference Hood and Skaar2012; Hennigar and McClung, Reference Hennigar and McClung2016). Four days post aerosol infection with APP, there was an increase in total iron binding capacity, iron in serum, and zinc in plasma (Humann-Ziehank et al., Reference Humann-Ziehank, Menzel, Roehrig, Schwert, Ganter and Hennig-Pauka2014). APP can sequester iron from host transferrin, heme, hemoglobin, and haptoglobin through the ferric uptake regulator (Fur) protein-dependent expression of genes encoding transferrin binding proteins (Tbps), heme binding protein A (HbpA), hemoglobin binding protein (HgbA), and a possible hemoglobin−haptoglobin binding protein (HpuB), respectively (reviewed in Chiers et al., Reference Chiers, De Waele, Pasmans, Ducatelle and Haesebrouck2010). APP acquires zinc through genes encoded by the znuABC operon (Yuan et al., Reference Yuan, Liao, You, Liu, Tan, Zheng, BinWang, Tian and Bei2014).
That iron and zinc are important for infection is clear from experimental studies in pigs. Firstly, surface-exposed APP iron and zinc-acquisition proteins are immunogenic (Goethe et al., Reference Goethe, Gonzales, Lindner and Gerlach2000; Liao et al., Reference Liao, Deng, Zhang, Zhou, Hu, Chen and Jin2009); secondly, attenuation of APP virulence in pigs can be achieved by mutation of genes involved in iron and zinc-acquisition, e.g. fur (Jacobsen et al., Reference Jacobsen, Gerstenberger, Gruber, Bossé, Langford, Hennig-Pauka, Meens and Gerlach2005) and znuA (Yuan et al., Reference Yuan, Liao, You, Liu, Tan, Zheng, BinWang, Tian and Bei2014). The data suggest that the ability of both host to sequester, and bacterium to obtain, iron and zinc contributes to disease susceptibility. That a prominent functional expression quantitative trait locus (eQTL) on Sus scrofa chromosome (SSC) 13, associated with resistance to APP infection, was found near the transferrin gene (Reiner et al., Reference Reiner, Dreher, Drungowski, Hoeltig, Bertsch, Selke, Willems, Gerlach, Probst, Tuemmler, Waldmann and Herwig2014b; see Genetics), and that among the six serovars investigated, the lowest expression of the three APP hpuB ORFs was found in serovar 3 (Klitgaard et al., Reference Klitgaard, Friis, Angen and Boye2010), one of the lowest virulent serovars (Rosendal et al., Reference Rosendal, Boyd and Gilbride1985), also supports a role for nutritional immunity in APP disease susceptibility.
Genetics
In the original MISTEACHING framework, only host genetics was considered under this category, and again will only be considered here. We could also have included discussion of pathogen genetics under this heading, but believe that bacterial genomic and phenotypic differences are best included under a new category – ‘Strain’ – for the reasons given in that section (see below).
That some breeds of pigs are more likely to die from bacterial lung infection is well documented, and suggests host genetics contributes to susceptibility/resistance to respiratory diseases (Jones, Reference Jones1969; Straw et al., Reference Straw, Neubauer and Leman1983). Vaccines are widely used in the control of APP (Loera-Muro and Angulo, Reference Loera-Muro and Angulo2018), thus pig lines bred to have higher immune responses offer a route for enhanced protection, and this has been investigated (Magnusson et al., Reference Magnusson, Bossé, Mallard, Rosendal and Wilkie1997). A commercial whole-cell killed (bacterin) vaccine was administered to Yorkshire pigs selected for high immune response (HIR) or low immune response (LIR). It was concluded, based on the antibody response to carbohydrate and LPS antigens, that HIR pigs had a greater immune response to the bacterin vaccine than LIR pigs, and that ‘multi-trait selective breeding may be useful in facilitating vaccine-based health management programs for livestock.’ More recently, a genomic study of replacement gilts identified quantatiative trait loci (QTLs) for antibody response to different serovars (1, 2, 3, 5, 7, 10, 12, and 13) of APP (Sanglard et al., Reference Sanglard, Mote, Willson, Harding, Plastow, Dekkers and Serão2020). Most QTLs identified were serovar-specific but one on SSC14 was associated with serovars 3, 5, 7 and 13, in a region associated with surface immunoglobulin IgM complexes. In total, genomic regions associated with the Ab response to the eight APP serovars were identified on 13 chromosomes. Several regions identified have been associated with reproductive traits in pigs, and the authors suggested that the QTLs could potentially be used for the improvement of resilience in commercial sows. That genetics is an important component of susceptibility/resistance to disease is also suggested by studies with transgenic pigs.
With a view to breeding pigs that are more resistant to APP, Hoeltig et al. formulated a new respiratory health score (RHS) and tested the susceptibility of four pig breeds (i.e. German Landrace, Piétrain, Hampshire, and Large White) to AP76, a serovar 7 strain of APP, delivered by aerosol (Hoeltig et al., Reference Hoeltig, Hennig-Pauka, Thies, Rehm, Beyerbach, Strutzberg-Minder, Gerlach and Waldmann2009). Based on the RHS, Hampshire and German Landrace pigs were the least and most susceptible to AP76 infection, respectively, confirming results from commercial farms (Jones, Reference Jones1969; Straw et al., Reference Straw, Neubauer and Leman1983). Follow-up studies to detect QTLs associated with susceptibility/resistance to AP76 on 170 Hampshire × German Landrace F2 animals, identified significant QTLs on SSCs 2, 6, 12, 13, 16, 17 and 18 that explained 6–22% of phenotypic variance (Reiner et al., Reference Reiner, Bertsch, Hoeltig, Selke, Willems, Gerlach, Tuemmler, Probst, Herwig, Drungowski and Waldmann2014a). One QTL on SSC2 reached significance on a genome-wide level for five associated phenotypic traits, and the genes IL-9 (encoding interleukin 9), and CD14 (encoding the cluster of differentiation 14 protein), known to be involved in the innate immune response to LPS, were considered candidates worthy of further investigation. A previous study by Gregersen et al. (Reference Gregersen, Sorensen, Christensen, Busch, Vingborg, Velander, Lund and Bendixen2010) on 7470 pigs from crosses between 12 Danish Duroc boars and 604 sows (Danish Landrace × Danish Large White) evaluated for dorsocaudal chronic pleuritis, which is a marker of pleuropneumonia (Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019), had identified QTLs on SSC2, 8, 12, 13, 14 and 18 as being associated with resistance to APP. Thus, QTLs on SSC8 and SSC14 were detected only by Gregersen et al. (Reference Gregersen, Sorensen, Christensen, Busch, Vingborg, Velander, Lund and Bendixen2010), SSCs 6, 16, and 17 by Reiner et al. (Reference Reiner, Bertsch, Hoeltig, Selke, Willems, Gerlach, Tuemmler, Probst, Herwig, Drungowski and Waldmann2014a), and SSCs 2, 12, 13, and 18 in both studies.
To prioritize candidate genes for fine mapping of gene variants, functional and transcriptomic analyses were carried out on the 50 most- and least-susceptible of the 170 pigs from the clinical QTL study (Reiner et al., Reference Reiner, Dreher, Drungowski, Hoeltig, Bertsch, Selke, Willems, Gerlach, Probst, Tuemmler, Waldmann and Herwig2014b). There were 171 differentially expressed genes between the two extremes of phenotype, and combined eQTL analyses (which identify associations between the expression of a specific gene and genotypes at different chromosomal locations) with network analyses and functional characterization, identified a functional hotspot on SSC13 which included 55 eQTLs. The most prominent of these 55 eQTLs, which explained 57% of total F2 variance, was a chromosomal region near the transferrin gene. APP can acquire iron for growth in the host by expression of Tbps (Gerlach et al., Reference Gerlach, Anderson, Potter, Klashinsky and Willson1992), thus the result is biologically plausible. While many candidate genes were discovered, the authors concluded that ‘Further research will be needed to prove or reject their causal role in susceptibility to A. pleuropneumoniae’.
A further fine-mapping study used next generation sequencing to genotype 58 German Landrace pigs with the most extreme phenotypes after aerosol delivery of AP76, and a genome-wide association study (resistant versus susceptible to infection) identified SNPs on SSC2, SSC12 and SSC15, which combined explained 52.8% of the variance (Nietfeld et al., Reference Nietfeld, Höltig, Willems, Valentin-Weigand, Wurmser, Waldmann, Fries and Reiner2020). The significant variants on SSC2, which explained 32.9% of the phenotypic variance, were mostly intronic or intergenic, but some were in the gene encoding f-spondin (or SPONDIN-1). In mice, this gene is responsible for maintaining circadian rhythms (Carrillo et al., Reference Carrillo, Su, Monavarfeshani and Fox2018), and is a negative regulator of bone mass (Palmer et al., Reference Palmer, Attur, Yang, Liu, Moon, Beier and Abramson2014), whose cellular expression in vascular smooth muscle cells is increased by LPS binding. Specifically, LPS binds to TLR4 activating the PI3K/Akt signaling pathway, inducing f-spondin expression and subsequent proinflammatory IL-6 production, to promote vascular smooth muscle cell migration (Lee et al., Reference Lee, Wu, Yeh and Kuo2016). IL-6 levels are known to dramatically increase during acute APP infection (Baarsch et al., Reference Baarsch, Scamurra, Burger, Foss, Maheswaran and Murtaugh1995). The SNP on SSC12 is an intron variant of the platelet and endothelial cell adhesion molecule 1 gene (PECAM 1), and explained 19.9% of phenotypic variance (Nietfeld et al., Reference Nietfeld, Höltig, Willems, Valentin-Weigand, Wurmser, Waldmann, Fries and Reiner2020). PECAM-1 (also called CD31) is a glycoprotein that is a member of the immunoglobulin gene (Ig) superfamily (Lertkiatmongkol et al., Reference Lertkiatmongkol, Paddock, Newman, Zhu, Thomas and Newman2016). In humans, PECAM-1 is expressed on the surface of granulocytes, monocytes, platelets, and endothelial cells where it functions to regulate vascular permeability and has a major role in leucocyte transendothelial migration (Privratsky et al., Reference Privratsky, Newman and Newman2010). The facilitation of leukocyte transendothelial migration is considered pro-inflammatory, while functions including dampening of leukocyte activation, suppression of pro-inflammatory cytokine production, and maintenance of endothelial barrier integrity are considered anti-inflammatory. In pigs, PECAM-1 has been identified as the receptor for Clostridium perfringens beta-toxin (Bruggisser et al., Reference Bruggisser, Tarek, Wyder, Muller, von Ballmoos, Witz, Enzmann, Deutsch, Engelhardt and Posthaus2020). PECAM-1 deficient mice were more sensitive to systemic administration of E. coli LPS than wild-type mice (Maas et al., Reference Maas, Stapleton, Bergom, Mattson, Newman and Newman2005). LPS administration was associated with excessive accumulation of macrophages and neutrophils in the lungs of PECAM-1-deficient mice, and correlated with a prolonged increase in lung pro-inflammatory cytokine (e.g. IL-6 and monocyte chemoattractant protein-1, and chemokine C-X-C motif ligand 1) levels. The diminishing accumulation of cytokine-producing leukocytes, rather than regulation of cytokine synthesis by leukocytes was proposed to explain the results (Privratsky et al., Reference Privratsky, Newman and Newman2010).
Infection with APP (or purified ApxI) results in apoptosis of porcine alveolar macrophages (Chien et al., Reference Chien, Chan, Chen, Wu, Liao, Chen, Lee, Yeh and Hsuan2009; Wang et al., Reference Wang, Qin, Ruidong, Liu, Zhang, Sun, Feng, Gu, Du, Han, Langford and Lei2015, Reference Wang, Qin, Zhang, Bao, Zhang, Che, Sun, Gu, Feng, Du, Han, Langford and Lei2016; Kim et al., Reference Kim, Oh, Bin Park and Yoo2019). Apoptotic cells are cleared by phagocytic cells involving ‘find me’ and ‘eat me’ signals (Grimsley and Ravichandran, Reference Grimsley and Ravichandran2003). PECAM-1 expression is known to be a ‘don't eat-me’ signal protecting non-apoptotic cells from clearance (Brown et al., Reference Brown, Heinisch, Ross, Shaw, Buckley and Savill2002). The above studies show that PECAM-1 has roles in inflammation and response to infection, and is a biologically plausible candidate worthy of further study in the context of APP infection. The variants on SSC15, associated with up to 18.5% of phenotypic variation, were in the COL4A4 region involved in the synthesis of type IV collagen, a backbone component of basement membranes. APP is known to bind to porcine lung-derived type IV (and types I, II, and III) collagen in vitro via an unidentified 60 kDa protein (Enríquez-Verdugo et al., Reference Enríquez-Verdugo, Guerrero, Serrano, Godinez, Rosales, Tenorio and de la Garza2004). It was speculated that the variants in the COL4A4 region on SSC15 may be part of mechanisms that restrict APP adhesion to pig lungs (Nietfeld et al., Reference Nietfeld, Höltig, Willems, Valentin-Weigand, Wurmser, Waldmann, Fries and Reiner2020).
Overexpression of porcine beta-defensin 2 (PBD-2) in transgenic pigs increased resistance (as adjudged by viable counts, lung severity scores, and histopathology) to APP intratracheal infection (Yang et al., Reference Yang, Cheng, Tan, Zhang, Liu, Zou, Zhang, Zhang, Deng, Yu, Hu, Li and Zhou2015). The same pigs were also more resistant to G. parasuis (Huang et al., Reference Huang, Yang, Wang, Huang, Tang, Zhang, Fang, Yu, Liu, Huang, Zhou and Li2020). For some infectious diseases, the contribution of bacterial and host genetic factors to susceptibility has been assessed by calculating the sibling risk ratio, which measures the increased risk of disease in siblings of affected cases compared with the risk in the general population. For N. meningitidis, approximately one third of susceptibility to disease was attributed to host genetics, and two thirds to other predominantly bacterial factors (Haralambous et al., Reference Haralambous, Weiss, Radalowicz, Hibberd, Booy and Levin2003). To our knowledge, no such analysis has been carried out with APP.
All of the above studies indicate that host genetic background is a contributor to resistance/susceptibility to APP infection, but that substantial further work is required to elucidate the identity of the specific genes/polymorphisms, and the underlying mechanisms involved, and whether such changes result in resistance/susceptibility to other pathogens as is the case with PBD-2 transgenic pigs. Also, before applying selective breeding, or even genome editing – which has recently been investigated for generation of pigs resistant to selected viral pathogens (Proudfoot et al., Reference Proudfoot, Lillico and Tait-Burkard2019), to alter specific genes for enhanced resistance to APP infection, it will be necessary to determine the effects of these mutations on other immune-related parameters (such as inflammation and stress) as well as overall health and production performance (Heuß et al., Reference Heuß, Pröll-Cornelissen, Neuhoff, Tholen and Große-Brinkhaus2019; Ballester et al., Reference Ballester, Ramayo-Caldas, González-Rodríguez, Pascual, Reixach, Díaz, Blanc, López-Serrano, Tibau and Quintanilla2020).
Strain
Here we propose that an additional category – Strain – be added to the MISTEACHING framework so that it becomes MISTEACHINGS. As originally formulated, the MISTEACHING model comprised only two categories that were clearly microbial-centric, i.e. Microbiome and Inoculum. APP has 19 serovars which are determined by surface carbohydrates, predominantly capsule (Bossé et al., Reference Bossé, Li, Fernandez Crespo, Lacouture, Gottschalk, Sarkozi, Fodor, Casas Amoribieta, Angen, Nedbalcova, Holden, Maskell, Tucker, Wren, Rycroft and Langford2018a; Stringer et al., Reference Stringer, Bossé, Lacouture, Gottschalk, Fodor, Angen, Velazquez, Penny, Lei, Langford and Li2021), and isolates express one or two of the ApxI-III toxins. ApxI is both strongly cytotoxic and haemolytic, ApxII is both weakly cytotoxic and hemolytic, and ApxIII is strongly cytotoxic (reviewed in Frey, Reference Frey1995). In general, there is a correlation between serovar and ApxI-III expression, although exceptions are increasingly being found (reviewed in Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). Isolates expressing ApxI in combination with ApxII (serovars 1, 5, 9, 11 and 16) are considered to be of high virulence, and those expressing ApxII and ApxIII (serovars 2, 4, 6, 8 and 15) of medium virulence. Isolates expressing only one of ApxI-III toxins are generally considered of low virulence (reviewed in Frey, Reference Frey1995; Gottschalk and Broes, Reference Gottschalk, Broes, Zimmerman, Karricker, Ramirez, Schwartz, Stevenson and Zhang2019). However, this is an oversimplification. For example, Jacobsen et al. found that serovar 2, 5, and 6 isolates were of equal virulence for pigs indicating that factors other than expression of ApxI and ApxII toxins are important (Jacobsen et al., Reference Jacobsen, Nielsen and Nielsen1996). While ApxI-III toxins are unquestionably virulence factors involved in induction of lesions (Chiers et al., Reference Chiers, De Waele, Pasmans, Ducatelle and Haesebrouck2010), mutants producing wild-type Apx toxins, but with mutations in non-Apx related genes, can be attenuated (Bossé et al., Reference Bossé, Janson, Sheehan, Beddek, Rycroft, Kroll and Langford2002; Chiers et al., Reference Chiers, De Waele, Pasmans, Ducatelle and Haesebrouck2010). In particular, signature tagged mutagenesis studies in pigs led to the discovery of many attenuating mutations unrelated to expression of Apx toxins (Fuller et al., Reference Fuller, Martin, Teel, Alaniz, Kennedy and Lowery2000; Sheehan et al., Reference Sheehan, Bossé, Beddek, Rycroft, Kroll and Langford2003). Encoded virulence factors were characterized as those involved in adhesion, acquisition of essential nutrients, avoiding host defense mechanisms, and persistence. An added complication is that the same mutation in different serovars may result in different phenotypic characteristics (Crispim et al., Reference Crispim, da Silva, Sanches, da Silva, Pereira, Rossi, Li, Terra, Vohra, Wren, Langford, Bossé and Bazzolli2020). Compared to many other pig pathogens, e.g. Streptococcus suis (Weinert et al., Reference Weinert, Chaudhuri, Wang, Peters, Corander, Jombart, Baig, Howell, Vehkala, Välimäki, Harris, Chieu, Van Vinh Chau, Campbell, Schultsz, Parkhill, Bentley, Langford, Rycroft, Wren, Farrar, Baker, Hoa, Holden, Tucker and Maskell2015), there is a comparative lack of APP whole genome sequences available. Such availability would enable many more strain-specific questions to be addressed. For example, with the closely related human pathogen, H. influenzae, it was shown that a subset of genes was essential for survival in animals co-infected with influenza virus rather than the bacterium alone (Wong et al., Reference Wong, Bernui, Shen and Akerley2013). APP is a primary pathogen of the PRDC and co-infections with other bacteria and viruses (including influenza) occur. No information exists, to our knowledge, on whether specific strains of APP, through their gene content, have a survival advantage during co-infections with other microorganisms.
APP has been classified as intermediately clonal, based on multilocus enzyme electrophoresis (Musser et al., Reference Musser, Rapp and Selander1987; Møller et al., Reference Møller, Nielsen, Andersen and Kilian1992) and amplified fragment-length polymorphism analysis (Kokotovic and Angen, Reference Kokotovic and Angen2007). To our knowledge there is no conclusive published evidence of specific pathotypes of APP, e.g. clades associated with diseases or asymptomatic carriage but not associated with disease; even low virulence serovars, such as 3, can cause disease (Rosendal et al., Reference Rosendal, Boyd and Gilbride1985). This contrasts with G. parasuis, also a pig pathogen within the family Pasteurellacaeae, which is highly diverse at the population level (Howell et al., Reference Howell, Weinert, Chaudhuri, Luan, Peters, Corander, Harris, Angen, Aragon, Bensaid, Williamson, Parkhill, Langford, Rycroft, Wren, Holden, Tucker and Maskell2014). In the case of G. parasuis, a pangenome study identified 48 genes associated with clinical disease, of which a subset of 10 was used to formulate a pathotyping PCR to aid herd surveillance and disease control (Howell et al., Reference Howell, Weinert, Peters, Wang, Hernandez-Garcia, Chaudhuri, Luan, Angen, Aragon, Williamson, Langford, Rycroft, Wren, Maskell and Tucker2017). Pathotypes have also been described for S. suis (Wileman et al., Reference Wileman, Weinert, Howell, Wang, Peters, Williamson, Wells, Langford, Rycroft, Wren, Maskell and Tucker2019), another bacterium that can cause disease in pigs. Similarly, for the human pathogen, N. meningitidis, some clonal complexes (such as CC11) are known to be disproportionately associated with invasive disease, while others have only been associated with carriage (Caugant and Brynildsrud, Reference Caugant and Brynildsrud2020).
Therefore, we suggest that the inoculum size is insufficiently descriptive of potential strain differences, especially where there is data suggesting pathotypes and/or specific clonal complexes being associated with disease. We, therefore, propose that the MISTEACHING framework be extended to MISTEACHINGS, with the addition of the Strain category, and that a research gap with APP is the availability of whole genome sequences.
Hypothesis generation and relation of MISTEACHINGS to other frameworks
In their original publication, Casadevall and Pirofski (Reference Casadevall and Pirofski2018) indicated that the MISTEACHING framework was applicable to any pathogen, but the emphasis was on those affecting humans. Here we show that the framework can be applied to veterinary pathogens, using APP as an exemplar. We additionally show, based on the discussion in the 12 sections above, that the MISTEACHINGS framework can be used to formulate hypotheses worthy of study to aid disease control (Table 1). A recent review (Sassu et al., Reference Sassu, Bossé, Tobias, Gottschalk, Langford and Hennig-Pauka2018) summarized gaps and challenges in APP research, identified under the DISease CONtrol TOOLS (DISCONTOOLS) framework (www.discontools.eu/), but the recommendations made were practical and applied, e.g. improvements in vaccines, diagnostics, and treatment, rather than hypothesis generating. The MISTEACHINGS framework can be considered as less rigid than DISCONTOOLS (which also includes a scoring system), but both identified different areas of future research to control APP, and therefore the two frameworks can be considered as highly complementary. Both frameworks indicate that susceptibility of pigs to APP is a complex host−pathogen interaction, with all of the MISTEACHINGS categories (with the exception of Sex) contributing to disease susceptibility. We also propose, for the reasons given above, that the MISTEACHING framework be extended to MISTEACHINGS to take into account strain variation, especially to accommodate high levels of genetic diversity within a species.
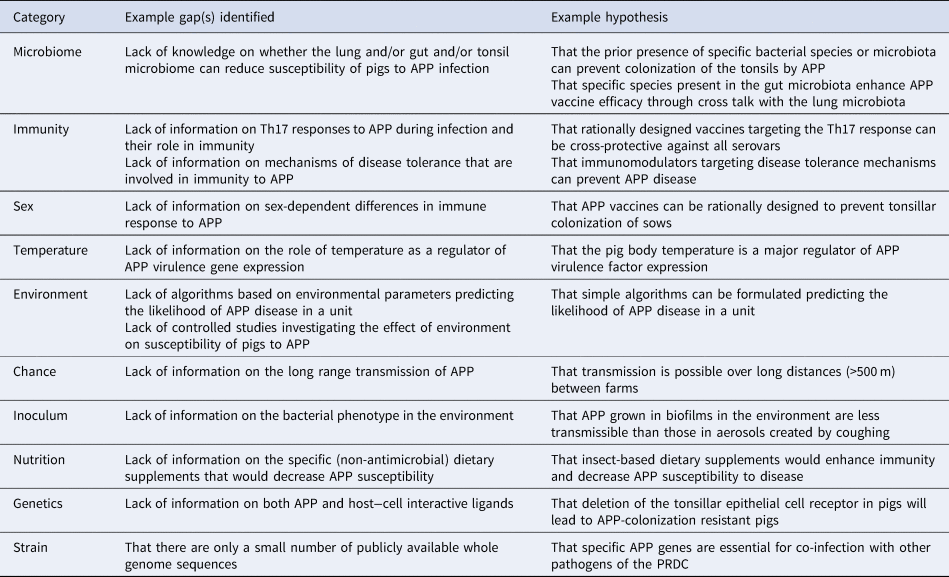
Table 1. Examples of gap(s) identified and hypotheses formulated from MISTEACHINGS analysis
The original MISTEACHING framework was host-centric, with only Microbiome and Inoculum being microbial-centric. Thus, the addition of Strain extends the microbe-centric component of the framework (Fig. 3). While there are many definitions of microbiomes, most involve the microbiota, genomic content, and local environment (Casadevall and Pirofski, Reference Casadevall and Pirofski2015; Berg et al., Reference Berg, Rybakova, Fischer, Cernava, Vergès, Charles, Chen, Cocolin, Eversole, Corral, Kazou, Kinkel, Lange, Lima, Loy, Macklin, Maguin, Mauchline, McClure, Mitter, Ryan, Sarand, Smidt, Schelkle, Roume, Kiran, Selvin, Souza, van Overbeek, Singh, Wagner, Walsh, Sessitsch and Schloter2020), and it could be considered as both a host and microbial factor, although it is clearly predominantly microbial in nature. With Temperature, Chance, and Nutrition, we have also considered both microbe and host-centric standpoints. However, we consider that the host centric elements are stronger than the microbe-centric elements and, therefore, categorized them as host-centric (Fig. 3). It must be acknowledged that the contribution of Chance is difficult to assess, and likely to have a minimal contribution to APP disease susceptibility.
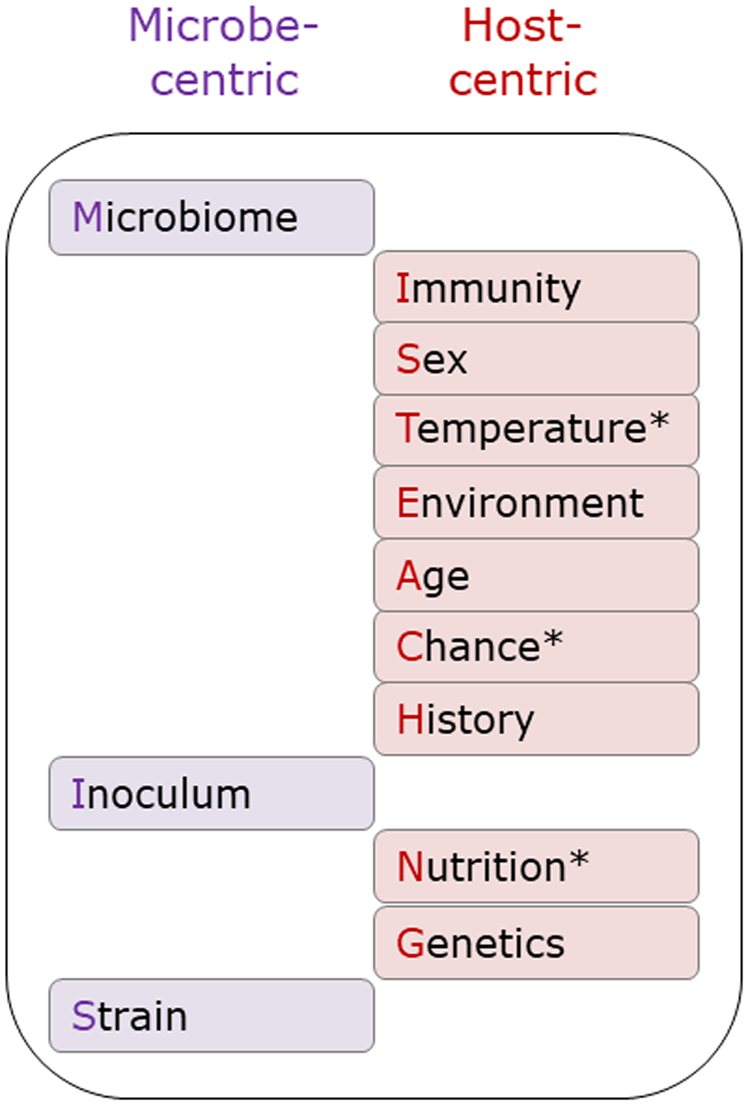
Fig. 3. Microbe and host-centric components of the MISTEACHINGS framework. * In the main text, Temperature, Chance, and Nutrition have been considered from both host and microbe perspectives. In our opinion, the host-centric elements of these three categories have a greater contribution to APP disease susceptibility than the microbe-centric element, hence their inclusion in the host-centric column.
APP is a damage response framework (DRF) class 2 pathogen
The MISTEACHING framework partly arose from previous work of Casadevall and Pirofski ‘struggling to find basic definitions of pathogenicity and virulence that incorporated the contributions of both the host and the pathogen’, which led to the DRF (Casadevall and Pirofski, Reference Casadevall and Pirofski1999). The basic tenets of the DRF are that: (1) microbial pathogenesis is an outcome of an interaction between a host and a microorganism; (2) the host-relevant outcome of the host–microorganism interaction is determined by the amount of damage to the host; and (3) host damage can result from microbial factors and/or the host response (Casadevall and Pirofski, Reference Casadevall and Pirofski2003). Under the DRF a pathogen is defined as ‘a microbe capable of causing host damage’, and damage as ‘disruptions in the normal homeostatic mechanisms of a host that alter the functioning of cells, tissues or organs; for microorganisms, disruptions in the normal mechanisms that enable host entry, replication and/or the ability to establish residence in a host.’ (Casadevall and Pirofski, Reference Casadevall and Pirofski1999). In the DRF, pathogens are separated into six classes. We propose that APP should be considered as a DRF Class 2 pathogen, i.e. it can cause damage either in hosts with weak immune responses or in the setting of normal host responses. The rationale is that APP causes significant damage in naïve animals (weak host response) and those bacteria that survive acute infection induce antibodies (strong host response) but can nevertheless continue to colonize the tonsils (Fig. 4). It should be noted that in the context of the DRF, colonization is defined as ‘a state of host–microorganism interaction that leads to a variable amount of host damage, from minimal to great, thereby reflecting host immune responses that have the capacity to eliminate the microorganism.’ (Casadevall and Pirofski, Reference Casadevall and Pirofski2003). APP also has two classic traits of Class 2 microorganisms, i.e. cause host damage by both pathogen (Apx toxins I-III) and host-mediated mechanisms, and that the immune response elicited by infection does not continue to damage the host once acute infection is resolved (Casadevall and Pirofski, Reference Casadevall and Pirofski1999).
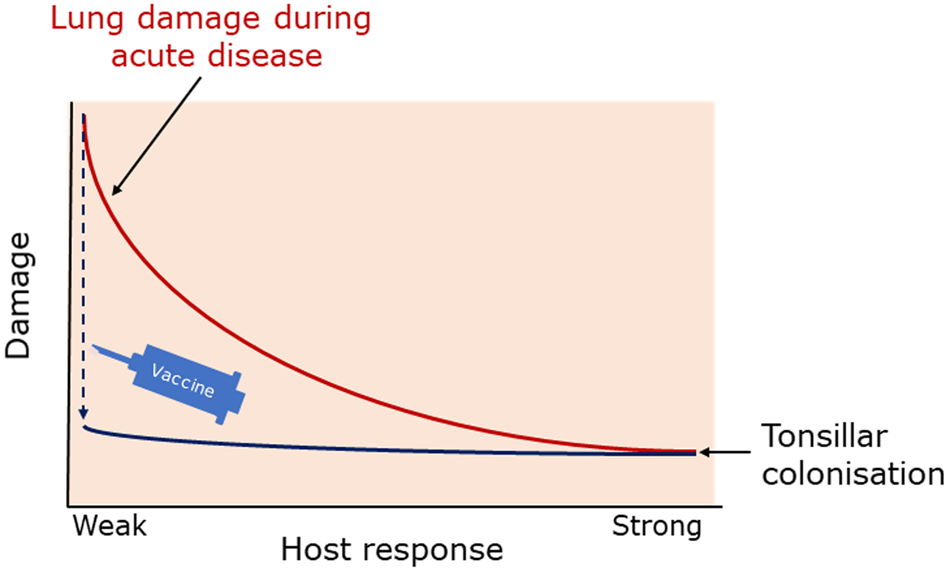
Fig. 4. APP is a Class 2 pathogen of the Damage Response Framework (Casadevall and Pirofski, Reference Casadevall and Pirofski1999). During acute infection of naïve animals, APP invade the lung and cause considerable damage, which may resolve in surviving animals due to actions of the host immune response. However, despite a strong humoral response, e.g. against the Apx toxins, APP can continue to persist and colonize the tonsils. In the DRF, colonization is considered to induce damage (even if it is minimal) hence the position of the curve in the Damage sector. Bacterin (whole cell) and Apx-based vaccines reduce or eliminate lung damage but do not prevent colonization, with the effect of flattening the damage response curve (blue line). Adapted from Casadevall and Pirofski (Reference Casadevall and Pirofski2003) with permission.
Conclusions
We have shown, using APP as an exemplar, that the MISTEACHINGS framework can be applied to a veterinary pathogen. We hope to stimulate others to use this framework to identify research gaps and to formulate hypotheses worthy of study in both veterinary research and teaching arenas with their veterinary pathogens, whether they be bacteria, fungi, viruses, or parasites.