Introduction
During recent decades, efforts have been made to describe global methane budgets accurately in order to understand the dynamics of the sinks and sources of this important greenhouse gas (Reference Adamsen and KingAdamsen and King, 1993; Reference KingKing, 1997; Reference Berestovskaya, Vasil’eva, Chestnykh and ZavarzinBerestovskaya and others, 2002, Reference Berestovskaya, Rusanov, Vasil’eva and Pimenov2005; Reference MastepanovMastepanov and others, 2008). Changes in the concentration of atmospheric methane, which is the most abundant organic trace gas in the atmosphere, play an important role in climate change due to its pronounced infrared absorption capacity. Since the industrial revolution the atmospheric CH4 concentration has increased as a consequence of imbalance between the biological and anthropogenic production of methane and its consumption in the environment (Reference Trotsenko and MurrellTrotsenko and Murrell, 2008). It has been estimated that approximately 30Tg of atmospheric CH4 per year are oxidized by aerobic methanotrophic bacteria in upland soils, accounting for about 6% of the global atmospheric CH4 sink (Reference Knief, Lipski and DunfieldKnief and others, 2003). Since the CH4 cycle is affected by feedback mechanisms from the ongoing global warming (Reference CallaghanCallaghan and others, 2004; Reference AnisimovAnisimov, 2007), the scientific community is now focusing on estimating potential climatic feedbacks from rapidly changing environments. An example of such an environment is recently deglaciated glacier forefields, where glacier recession exposes previously subglacial ecosystems to proglacial climatic conditions. As a consequence, changes in microbial community composition and activity may alter net respiration rates and hence affect the consumption or emission of greenhouse gases such as CO2, CH4 and N2O. In recent years a growing number of publications have focused on issues concerning glacier forefield ecology (Reference Sigler and ZeyerSigler and Zeyer, 2002; Reference Sigler, Crivii and ZeyerSigler and others, 2002; Reference Tscherko, Rustemeier, Richter, Wanek and KandelerTscherko and others, 2003; Reference Bekku, Nakatsubo, Kume and KoizumiBekku and others, 2004; Reference Kaštovská, Elster, Stibal and ŠantréčkováKaštovská and others, 2005; Reference Kandeler, Deiglmayr, Tscherko, Bru and PhilippotKandeler and others, 2006; Reference BardgettBardgett and others, 2007; Reference Hämmerli, Waldhuber, Miniaci, Zeyer and BungeHämmerli and others, 2007; Reference NemergutNemergut and others, 2007; Reference Yoshitake, Uchida, Koizumi and NakatsuboYoshitake and others, 2007; Reference SchmidtSchmidt and others, 2008; Reference Duc, Noll, Meier, Bürgmann and ZeyerDuc and others, 2009; Reference Lazzaro, Abegg and ZeyerLazzaro and others, 2009; Reference Strauss, Ruhland and DayStrauss and others, 2009). Many of these studies have identified changes in microbial community structures along chronosequence profiles. However, a gap still exists in knowledge about whether glacier recession causes net microbial consumption or emission of CH4 and how significant these changes are to the global CH4 budget. In particular, little attention has been paid to glacier forefields in Greenland, where the recession of glaciers is occurring very fast (Reference Yde and KnudsenYde and Knudsen, 2007; Reference Mernild, Kane, Hansen, Jakobsen, Hasholt and KnudsenMernild and others, 2008). In this paper we present and discuss data on methane fluxes and the underlying methanotrophic microbial community which were obtained during a field campaign to the Mittivakkat valley, Greenland, in August 2009.
Study Area
The research area is located in the Mittivakkat valley (65842’ N, 31848’ W), Ammassalik Island, southeast Greenland (Fig. 1). The valley was covered by Mittivakkat Glacier, an outlet glacier descending from a local ice cap, until the end of the Little Ice Age (LIA). It is not known when the glacier started to recede from the LIA moraine, but it was probably between AD1850 and 1900. It has been documented that since 1933 the glacier terminus has receded approximately 1.3 km to its current position (Reference FristrupFristrup, 1970; Reference Knudsen, Nønberg, Yde, Hasholt and HeinemeierKnudsen and others, 2008). The overall deglaciated valley geomorphology consists of a frontal dump moraine that marks the maximum extent of the LIA and a classic set of successively incised glaciofluvial outwash surfaces, where the highest surface is oldest and the lowest surface is used by the proglacial river today. This clear landform chronology makes the Mittivakkat valley an ideal site for investigating glacier forefield ecology.

Fig. 1. Location map of the Mittivakkat valley and the proglacial zone with the positions of the flux chamber sites.
The climate is Low Arctic, with a mean annual precipitation of 984 mm (AD1961–1990) and a mean annual air temperature (MAAT) of –1.7°C (AD 1961–90), but in the last decade the MAAT has increased to about 0.0°C (Reference CappelenCappelen, 2009a). The mean monthly air temperature (AD 1961–90) ranges from –8.1°C in March to 6.4°C in July (Reference CappelenCappelen, 2009b). There is no contemporary permafrost and the LIA moraine is not ice-cored. The vegetation cover is sparse and dominated by a dwarf shrub heath habitat with dwarf birch, greyleaf willow and fell-field communities based on cushion plants (Reference Jakobsen, Fredskild and PedersenJakobsen and others, 2008) as well as an abundance of lichens (Reference HansenHansen, 2010). The geology of Ammassalik Island mainly consists of Archean granite gneiss (Reference Bridgewater, Escher and WattBridgwater, 1976).
Methods
In August 2009, a total of 12 sites was studied along a proglacial transect from the LIA moraine to the present glacier terminus (Fig. 1). Sites C1A–C1D were located across the LIA moraine, with C1A and C1B on the distal side, C1C on the top and C1D on the proximal side; sites C2–C4 were situated on glaciofluvial outwash surfaces inside the LIA moraine, where C2 was oldest and C4 was youngest; site C5 was located on a small moraine ridge that was younger than C4 and deposited some years before 1933 (Reference FristrupFristrup, 1970); site GF1 was on top of a moraine ridge, which was deglaciated a few years after 1958 (Reference FristrupFristrup, 1970); site GF2 was situated on glaciofluvial sediments within a tributary valley and was exposed in the 1980s; site GF3 consisted of glaciofluvial sediments and was deglaciated during the summer of 2004; and site GF4 was located about 1 m from the glacier terminus in August 2009 and had been deglaciated within the previous year. The reason for the intensive sampling of the LIA moraine was that we expected differences in microclimatic environments, microscale slope processes and soil development on the distal side, top and proximal side, respectively.
Methane flux analysis
Methane fluxes were determined by the static chamber technique (Reference Priemé and ChristensenPrieme and Christensen, 1997; Reference SmithSmith and others, 2002; Reference Berestovskaya, Rusanov, Vasil’eva and PimenovBerestovskaya and others, 2005) combined with flame ionization gas chromatography. The chambers were 25 cm long polyvinylchloride cylinders with an internal diameter of 9.4 cm, which were inserted 10 cm into the sediment to give a final headspace volume of ∼1 L. The chambers were left open to the atmosphere for approximately 1 day before the beginning of the experiment. Samples of air at the same level where the chambers were placed were collected before the beginning of the experiment to determine initial methane atmospheric concentrations at each site. Sampling ports (9 mm diameter) on every chamber were then sealed with a rubber stopper to ensure gas-tight conditions throughout the experiment. Gas samples were withdrawn with a syringe from the headspace of the chambers and stored in 5mL evacuated Exetainer vials (Labco Ltd, Buckinghamshire, UK) containing 1 mL of 1 M NaOH. Gas samples were taken at 24 hour intervals during a period of up to 5 days. Three gas samples at each time of analysis were taken for the four chambers on the LIA terminal moraine ridge (C1A, C1B, C1C and C1D), while two gas samples were taken for the rest of the chambers in the chronosequence. After sampling gas from the headspace of the chambers, the withdrawn volume was compensated with the same volume in atmospheric air to maintain pressure conditions within the chamber. Analysed methane concentrations were corrected for this addition of methane. Analysis of the CH4 was performed by gas chromatography on an SRI 310C GC (SRI, Torrance, CA, USA) with a 3 ft × 1/8 in (∼0.91 m × 3.18 mm) silica-gel packed column (column i.d. 8600-PK1A) using helium as the carrier gas. Oven temperature was 40°C and the temperature in the field ionization detector was 155°C. Gas samples (500 μL) were injected into the GC system and analysed by simultaneous integration of the peaks in the chromatograph with the Peak Simple 2000 software (SRI, Torrance, CA, USA). Based on this set-up it was possible to reliably detect CH4 concentrations down to 0.2 ppm. Standards were run in the system before and after the measurements to verify their quality and perform calibration.
The oxidation rate of atmospheric methane was estimated from first-order decreases in headspace methane concentrations based on:
where C 0 is initial CH4 concentration, k is a first-order rate constant for CH4 consumption at approximately atmospheric concentrations (Ishizuka and others, Reference Ishizuka2009) and t is time. We followed changes in methane concentrations for a time period of 72 hours, because we were confident that the measurements based on this time interval could not arise from analytical artefacts.
pH
The pH analysis was performed in distilled water and 1 M KCl (two subsamples from each site) with a pHM 64 Research pH meter (Radiometer, Copenhagen, Denmark) to determine the actual and potential acidity of the soil sample, respectively. 10g of sediment sample and 10g of fluid (distilled water or KCl) were weighed into 20 mL plastic beakers and stirred four times during 30 min, and left afterwards for 30 min without stirring. The glass electrode was adjusted in buffer solutions and set to the temperature of the fluid before it was lowered into the sample to conduct the measurement (Reference Nørnberg and DalsgaardNørnberg and Dalsgaard, 2005).
Molecular analysis based on pmoA genes
Soil DNA extraction
Sediment subsamples (approximately 0.5 g) from each study site were used for DNA extraction and purification with the Fast DNA Spin Kit for Soil according to the manufacturer’s instructions (Bio101, La Jolla, CA, USA).
Genetic fingerprinting: denaturing gradient gel electrophoresis
In order to assess the presence of methanotrophs, a prior analysis based on polymerase chain reaction (PCR) was performed, using the primer set A189F-A650R targeting the pmoA functional gene sequence (Reference Bourne, McDonald and MurrellBourne and others, 2001). This primer set encodes one of the subunits of the particulate methane monooxygenase (pMMO), an enzyme found universally in most methanotrophs (for exceptions see Reference DedyshDedysh and others, 2000). After ensuring that the PCR product had the correct size, aGC clamp was added to the forward primer (GC-A189F) for further analysis with denaturing gradient gel electrophoresis (DGGE). The analysis was based on two equal gels (0.75 mm × 16mm × 16mm) of 8% w/v acrylamide in a denaturant gradient from 30% to 70%. The low denaturant was based on a mixture of 20% w/v urea and formamide in 8% w/v acrylamide, while a mixture of 80% w/v urea and formamide in 8% w/v acrylamide was used for the high denaturant. The same PCR products were loaded on both gels, with slight differences in loaded volume on each gel (gel 2 had 1 or 2 μL higher load of PCR product compared with gel 1). The gels were run in 1 × TAE buffer (40 mM Tris-HCl, 20 mM acetic acid, 1 mM EDTA, pH 8.3) at 60°C and 100 V for 15 hours using the DCode Universal Mutation Detection System electrophoresis apparatus (BioRad, Hercules, CA, USA) connected to a PowerPac 300 power supply (BioRad, Hercules, CA, USA). The gels were stained in a bath containing 200 mL of 1 × TAE buffer and 10 μL of DNA staining dye Sybr Gold (Invitrogen, Carlsbad, CA, USA) for 60 min. Finally, results were visualized with UV translumination on the GelDoc 2000 system (BioRad, Hercules, CA, USA) and photographed for further analysis.
Bands from both DGGE gels were excised for further analysis based on sequencing. The sequencing was conducted by the commercial company Macrogen (Korea).
Band analysis of the DGGE images
Band patterns from both gels were analysed and processed with the Gel Doc 2000 gel documentation system and the Quantity One (version 4.2.1) software (BioRad, Hercules, CA, USA) based on peak densities (i.e. the intensity value of a band’s peak) of each band as a main parameter to compare between lanes and bands. Band analysis was performed with precisely the same settings (sensitivity, noise filter, minimum density, etc.) and background substraction (based on the rolling disk method) on both gels to compare the band retrieval depending on the differences in sample load between gels. Sensitivity (i.e. the minimum signal intensity defined as a band) was determined with an absolute value of four, to avoid erroneous detection of bands. We produced two identical gels with different loads to ensure consistent sequencing results and to determine the effect of different volumes on the band patterns.
In addition, DGGE profiles were analysed for diversity based on the Shannon index (H ’) according to:
where ni corresponds to the peak density of each detected band and N represents the total peak density of all bands in one lane, giving the theoretical proportional abundance of different pmoA sequences within the different lanes (Reference Sigler and ZeyerSigler and Zeyer, 2002; Reference Trevors, Kevan and TamTrevors and others, 2010).
DGGE band sequence analysis
The sequences that were retrieved successfully from the DGGE bands were analysed using the NCBI BLAST tool with the NCBI nt database as reference (Reference AltschulAltschul and others, 1997). They were grouped according to their affiliation to sequences in the database.
Results
Methane flux
In situ net methane oxidation rates were detected in 7 of the 12 chambers and ranged between 0.14 and 0.76 μgCH4 h−1 m−2 ±0.02–0.05 μgCH4h−1 m−2 (Fig. 2). Four chambers did not show significant changes in CH4 headspace concentrations over the course of the experiment (C4, GF1, GF2 and GF3). An efflux of CH4 (net CH4 production) was observed in the chamber located ∼ 1 m from the glacier front (GF4). This site represents a deglaciation that took place between the summers of 2008 and 2009.

Fig. 2. Net flux rates calculated from headspace methane concentrations in the flux chambers over time, along the Mittivakkat valley chronosequence. Negative values represent net methane consumption and positive values represent net methane production.
The highest CH4 oxidation rate was determined on top of the LIA moraine ridge (site C1C) and reached 0.76 μgCH4 h−1 m−2. The variability in the rates of CH4 oxidation across the LIA moraine (sites C1A-C1D) showed that the CH4 oxidation rate increased towards the top of the moraine ridge. The reasons for this have yet to be determined, but it is likely that the stage of soil development and stability are important controls. As a general pattern, it was observed that the CH4 oxidation potential tends to decrease along the chronosequence towards the glacier front.
pH
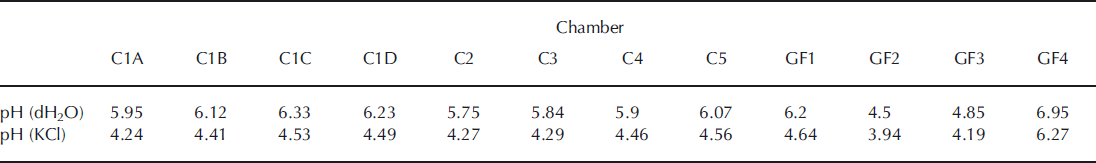
The pH values indicate an acidic environment varying in the range 3.9–4.6 in 11 of the 12 sampling sites, based on values obtained from 1 M KCl (Table 1). A relatively higher pH value (6.3) was found near the glacier terminus at site GF4. Unpublished pH measurements in glacier meltwater at Mittivakkat Glacier showed values between 6.4 and 7.3 in August (data not shown). This indicates that glacial meltwater most likely infiltrates the near-marginal glaciofluvial sediments at site GF4.
Table 1. pH values in the top soil at the sampling sites along the Mittivakkat valley chronosequence
Methanotrophic diversity
The DGGE patterns indicate a shift in diversity of pmoA gene sequences along the chronosequence. Subsamples obtained from top soil of the LIA terminal moraine ridge presented almost identical diversity patterns (Fig. 3a and b), demonstrating the homogeneous distribution of the methanotrophic microbial community within the same geomorphological unit. However, the patterns observed along the entire chronosequence changed gradually towards the youngest part.

Fig. 3. DGGE images of the Mittivakkat valley chronosequence. Numbers shown on the gels correspond to the band indicated by the triangle, which was extracted for further analysis (sequencing). Band numbers 1.1–1.32 (total of 32 bands) and 2.1–2.24 (total of 24 bands) refer to (a) the first and (b) the second gel. The amount of sample loaded in the gel shown in (a) ranged from 7 to 11 μL depending on the strength of the PCR product, while the gel in (b) contained 1–2 μL extra sample load. Both gels have the same 30–70% denaturant gradient.
No pmoA genes could be retrieved from sites GF2 and GF4; therefore, these samples were not included in the DGGE analysis.
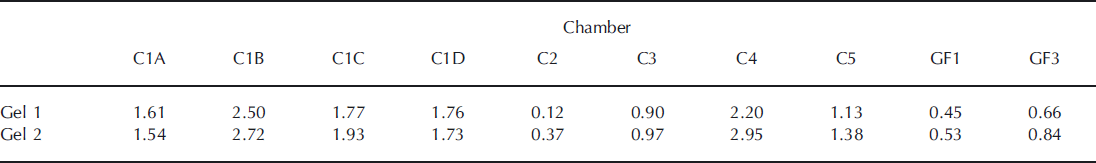
Based on the number of bands detected on the DGGE images, samples from sites C1B and C4 contained the highest diversity of all studied sites. These patterns were consistent, since they were retrieved on both gels. In general, the Shannon index of diversity increased in the second gel (Table 2), where the volume of sample was larger and more bands in almost all lanes were detected.
Table 2. Values for the Shannon diversity index (H’) in both gels
A comparison of the 55 out of 56 sequences that could be retrieved successfully from the DGGE bands with sequences stored in the NCBI nt database confirmed that all sequences were pmoA gene sequences. With the exception of a single sequence all sequences were closely affiliated with sequences that we recently obtained by cloning and sequencing from soil samples collected at the LIA moraine site and C5 in summer 2008. Of the 54 sequences, 28 fell into cluster 4, 19 into cluster 3, 6 into cluster 2, and 1 into cluster 1 (Table 3). These clusters fell into a group of high-affinity methanotrophs within the α-subdivision of Proteobacteria.
Table 3. Affiliation of sequences retrieved from DGGE bands with sequences stored in the GenBank database. All sequences affiliated with sequences of high-affinity type II methanotrophs within the α -subdivision of Proteobacteria

The applied methods demonstrate that the amount of PCR product loaded on the DGGE gel positively affected the number and the intensity of single bands in the gel upon staining, enhancing the importance of considering this factor relevant for the achievement of a comprehensive DGGE result.
Discussion
Methane Flux
Methane uptake rates in soils vary in a relatively wide range depending on ecosystem characteristics (Reference KingKing, 1997; Reference SmithSmith and others, 2002). A study from Reference Knief, Lipski and DunfieldKnief and others (2003) shows that methanotrophic upland soil communities (i.e. well drained and oxic) are able to consume methane at atmospheric concentrations. They argue that aerobic methanotrophic communities inhabiting these environments present high affinity for methane and should therefore be considered as highly specialized oligotrophs. In this study we are able to show that communities with equivalent properties inhabit a deglaciated forefield in southeast Greenland. Like the upland soil community studied by Reference Knief, Lipski and DunfieldKnief and others (2003), the glacier forefield community consumes methane at atmospheric levels. This is, to our knowledge, the first report of in situ CH4 flux measurements in this habitat type.
Methane flux studies based on flux chambers may present some methodic limitations. During an experiment, CH4 depletion in the headspace of the chamber may cause a reduction in methanotrophic activity compared with the surrounding soil that is exposed to ambient CH4, and lateral CH4 diffusion could supplement the soil methane supply (Reference Von Fischer, Butters, Duchateau, Thelwell and Sillervon Fischer and others, 2009). Also, increased temperature or moisture inside the chamber may affect the observed methanotrophic potential. Despite these limitations of the experimental set-up, we are convinced that the observed trends are real and reflect the situation in situ. However, more detailed studies need to be conducted in the future to evaluate the quality of the results.
Consumption rates at the Mittivakkat Glacier forefield are low compared with those measured in forest soils (Reference IshizukaIshizuka and others, 2009; Reference Jang, Lee, Hong and KangJang and others, 2009), tundra environments (Reference Whalen and ReeburghWhalen and Reeburgh, 1990) or landfill areas (Reference Hanson and HansonHanson and Hanson, 1996) and lie at the low end of rates reported so far (Table 4).
Table 4. Rates of atmospheric methane consumption measured in different low-rate environments, based on flux chamber experiments

The data presented in this study demonstrate that glacier forefields constitute potential sinks of CH4 during late stages after glacial recession (after 80–150 years of exposure). Even though the potential of these areas as a methane sink is modest, resembling the capacity of, for instance, arable soil (Reference Dobbie, Smith, Priemé, Christensen, Degόrska and OrlanskiDobbie and others, 1996) to oxidize CH4, it is necessary to take them into account as part of the CH4 sink budget in upland soils. The need to include this type of environment is also supported by the fact that areas representing deglaciated forefields are increasing in size worldwide as a result of enhanced glacier recession.
Besides methanotrophy, methanogenesis is a relevant process in glacier forefields. Based on the presented net flux data, net CH4 production and release to the atmosphere was observed in the chamber located immediately in front of the glacier terminus (GF4). This observation of methanogenesis in recently exposed material is in accordance with findings reported by Reference Wadham, Tranter, Tulaczyk and SharpWadham and others (2008), who presented evidence for subglacial methanogenesis.
We suggest that the observed shift in net flux CH4 from being methane-releasing at the glacier front to becoming methane-consuming in the older parts of the chronosequence is most likely a result of a changing availability of oxygen. The recently exposed sediments at the glacier front are most likely anoxic and thus support methanogenic Archaea, whereas the older sediments in the forefield area are well oxygenated and thus support a methanotrophic microbial community. Archaeal clone libraries from soil collected at site C1A (outermost terminal moraine) and the glacial front location GF4 support this argument (data not shown). While the GF4 Archaea community was dominated by methanogen-related 16S rDNA clones, the C1A library was free of methanogen-related 16S rDNA clones but contained a diverse archaeal community.
The pH at the different sampling locations deserves special consideration because it is an important environmental factor that has a strong influence on the distribution and activity of methanotrophs and methanogens across the forefield. Since the Mittivakkat Glacier forefield is only poorly buffered, shifts in pH are likely to take place and pH might therefore have had a significant influence on the shaping of the methanotrophic soil community during glacier recession since the LIA. Hitherto, only view reports on methanotrophs from acidic Arctic environments have been published (Reference DedyshDedysh and others, 2000). Thus, our report adds to the sparse number of observations on the activity and diversity of methanotrophs in this habitat type. We hope to follow up on this issue with additional studies in the near future.
Overall, we hypothesize that as redox conditions, access to oxygen and pH change, the microbial communities involved in CH4 cycling are changing accordingly as long as types that are able to meet the environmental constraints of the specific systems are present within the pool of methanotrophic bacterial strains. The interactions between the environmental conditions and the responsive microbial community are ultimately reflected in the net methane fluxes.
Methanotrophic diversity
A successful retrieval of pmoA gene sequences in all samples from the chronosequence, with the exception of two (GF2 and GF4), was achieved. However, the detection of a particular pmoA gene in a soil does not necessarily imply that the respective methanotroph is physiologically active in the soil (Reference Knief, Lipski and DunfieldKnief and others, 2003). In a previous study we observed differences between activity and presence of methane oxidizers in samples from the Mittivakkat Glacier forefield (unpublished information). In this study, we successfully retrieved pmoA genes from two of the samples where no activity could be detected previously (C4 and GF1), indicating that discrepancies exist between measurable activity and detectable presence. This may be due to the amplification of genetic material by PCR, which may render even small populations visible, the activity of which may not be observable due to the detection limits from the GC method.
With the purpose of performing diversity analyses, individual PCR products from all studied sites were separated by DGGE resulting in different band patterns, of which each band was theoretically representing a predominant member in the targeted microbial community, in this case high-affinity methanotrophs. Limitations of different molecular techniques such as DGGE have been described previously (Reference Muyzer and SmallaMuyzer and Smalla, 1998). In the case of DGGE analysis, it has been demonstrated that it is not always possible to separate DNA fragments with a certain sequence variation, which has been the case for DGGE diversity studies on methanotrophic bacteria based on 16S rDNA fragments (Reference VallaeysVallaeys and others, 1996). However, in this study we focus on the functional gene pmoA. In a previous diversity study, some difficulties were encountered when extraction and sequencing of the extracted DGGE bands were performed, probably due to a poor separation between DGGE bands. Cloning was therefore required. In the present study, a successful band separation in the gel made it possible to excise and sequence the obtained bands and thereby obtain pmoA gene sequences and compare them with previous results. Interestingly, all retrieved sequences fell into the clusters described in a previous study (unpublished information) and which affiliate with high-affinity type II methanotrophs. As was also the case when clone libraries were investigated, the majority of DGGE band sequences were closely affiliated with cluster 4 sequences. These independent observations confirm that the dominant methanotroph type in the deglaciated Mittivakkat valley belongs to a community of methanotrophs with a restricted diversity. A restricted diversity is also reported by Reference Liebner, Rublack, Stuehrmann and WagnerLiebner and others (2009) in a study of methanotrophic community composition in Arctic permafrost. The authors explain the limited diversity by the fact that these organisms have to cope with a combination of environmental extremes that only a limited number of methanotrophs can respond to. This argument may also be applicable when explaining the restricted diversity in the Mittivakkat Glacier forefield. In addition, restricted diversity was also reported from other extreme environments such as peat bogs and acidic forest soil (Reference McDonald, Hall, Pickup and MurrellMcDonald and others, 1996; Reference McDonald and MurrellMcDonald and Murrell, 1997; Reference RadajewskiRadajewski and others, 2002)
Our observation of the exclusive presence of high-affinity type II methanotrophs is in contrast to reports of methanotrophic diversity in Northern Hemisphere permafrost methanotroph communities (Reference Liebner and WagnerLiebner and Wagner, 2007; Reference Liebner, Rublack, Stuehrmann and WagnerLiebner and others, 2009; Reference Martineau, Whyte and GreerMartineau and others, 2010). These studies report on the dominant retrieval of low-affinity type I methanotrophs in the investigated permafrost soil. This difference in community composition is most likely due to the fact that the methane concentrations in the active layer of the permafrost soil are much higher as a result of indigenous methane production compared with the forefield sediments that we investigated and that glacier forefields receive methane exclusively from the atmosphere. A similar dominance of type II methanotrophs was also reported from other habitats such as different forest and grassland soils that, like the Mittivakkat Glacier forefield, receive methane only at atmospheric concentrations (Reference Ricke, Kube, Nakagawa, Erkel, Reinhardt and LiesackRicke and others, 2005; Reference Lau, Ahmad, Steudler and CavanaughLau and others, 2007).
Conclusion
Our results demonstrate clearly that recently deglaciated areas ultimately may act as a net sink for atmospheric CH4. We recommend further studies of the proglacial zone of receding glaciers to address more accurately the dynamics of net CH4 fluxes in these highly dynamic environments.
In addition, sites like the Mittivakkat valley are excellent environments in which to study the in situ colonization by pioneer microbial communities and to study the community shift upon transition from anoxic methanogenic subglacial conditions to oxic methanotrophic proglacial conditions. Overall, the Mittivakkat Glacier forefield represents an excellent natural laboratory in which the important consequences of climate-induced glacier retreat can be followed in situ.
Acknowledgements
This research was funded by the Commission for Scientific Research in Greenland (KVUG – project No. 495229), Carlsbergfondet and the Faculty of Science, University of Aarhus. We thank N.T. Knudsen, T. Wiegers, S.M. Kristiansen and E.N. Bak for field assistance and S. Flury for inspiring discussions. T. Wiegers is also acknowledged for her valuable assistance with molecular analyses and fieldwork. R. Nielsen is acknowledged for map design. We also thank B. Eriksen, B. Rasmussen and M. Sloth for their contribution with pH analyses. We are grateful to the Department of Geography and Geology (IGG), University of Copenhagen, for allowing us to use the Sermilik Research Station. This is publication No. A309 from the Bjerknes Centre for Climate Research.









