Introduction
There is a remarkable need for progress in the practice of clinical psychiatry. Mental illness and substance use disorders are the leading cause of disability worldwide with major depressive disorder (MDD) the commonest cause,Reference Whiteford, Degenhardt, Rehm, Baxter, Ferrari and Erskine1,2 suicide is a leading cause of death in young adults,2 and severe and enduring mental illness is associated with a reduction in lifespan of about a decade.Reference Chang, Hayes, Perera, Broadbent, Fernandes and Lee3 However, it is widely recognised that clinical practice in psychiatry has not fundamentally changed for over half a century.Reference Stephan, Bach, Fletcher, Flint, Frank and Friston4,Reference Stephan, Binder, Breakspear, Dayan, Johnstone and Meyer-Lindenberg5
During this time neuroscience has developed a range of sophisticated techniques allowing brain structure and function to be non-invasively investigated in unprecedented ways.Reference Shenton and Turetski6 Magnetic resonance imaging (MRI) allows non-invasive mapping of brain structure and function (fMRI), magnetoencephalography allows mapping of magnetic fields occurring naturally in the brain complementing electroencephalography (EEG) and positron emission tomography allows mapping of neurotransmitters. Countless neuroimaging studies have reported statistically significant differences between groups of patients and healthy controls and findings from ever larger molecular genetics studies are being reported. However, despite these incredible scientific and methodological advances, no progress has been made in applying neuroscience pragmatically to psychiatry.
Responding to this need, the goal of pragmatic psychiatric neuroscienceReference Paulus, Huang, Harle, Redish and Gordon7 is to develop neuroscience-based objective quantitative markers that aid clinical decision-making and have implications for individual patients, for example by objectifying diagnosis, quantifying prognosis, supporting treatment selection and yielding objective severity markers for disease monitoring. For example, it can often take months or years of trying different antidepressants to find one that is effective (if any) for a particular patient. There is good evidence for some antidepressants being more effective and tolerable than others,Reference Cipriani, Furukawa, Salanti, Chaimani, Atkinson and Ogawa8 yet different patients respond to different antidepressants without a clinically obvious pattern.Reference Rush, Kilner, Fava, Wisniewski, Warden and Nierenberg9 We would like to be able to say, ‘Mrs Smith, your depression test has come back and with this particular antidepressant, you have a 90% chance of being symptom free in 6 weeks’.Reference Paulus10
Traditional group-level statistical significance framework
There are various reasons for the limited impact of neuroscience on clinical psychiatry.
First, reproducibility of research findings has been a problem.Reference Collaboration11 Only recently have very large sample sizes become available to provide definitive group-level results: for example the ENIGMA Consortium recently reported a meta-analysis of structural brain scans from 8921 patients with MDD and controls from sites worldwide.Reference Schmaal, Veltman, van Erp, Sämann, Frodl and Jahanshad12 Patients had significantly lower hippocampal volumes with the authors concluding their study robustly identified hippocampal volume reductions in MDD.Reference Schmaal, Veltman, van Erp, Sämann, Frodl and Jahanshad12 This is a definitive result, however, it is not clinically useful for individual patient management, as will be discussed. Far larger psychiatric genetics studies typically failed to identify significant findings, although a recent study with 480 359 participants with MDD and controls did report significant findings.Reference Wray, Ripke, Mattheisen, Trzaskowski, Byrne and Abdellaoui13 Very small genetics effect sizes are unlikely to be useful for decision-making with individual patients; a different statistical framework is required.Reference Lo, Chernoff, Zheng and Lo14,Reference Lo, Chernoff, Zheng and Lo15
Second, psychiatric disorders are fundamentally multivariate constructs. When taking a psychiatric history and conducting a mental state examination, multiple symptom and social areas are explored before decision-making such as diagnosis and treatment selection. Therefore it should not be surprising that a single univariate measure, such as a single interview question, hippocampal volume or a single genetic measure is insufficient to substitute for this. Indeed, the National Institute of Mental Health, Research Domain Criteria (RDoC), currently has five domains of human function within which there are multiple ‘units of analyses’.Reference Insel, Cuthbert, Garvey, Heinssen, Pine and Quinn16,Reference Kendler17
The main problem with a group-level statistical significance framework is that it only allows inferences about group differences (for example average hippocampal volume in MDD versus controls) and not individual patients. Using this approach with psychiatric disorders, there is invariably substantial overlap in the distribution of a single variable for individual patients and controls, meaning that if such a measure is used in an attempt to make a clinically relevant prediction for an individual patient, the predictive accuracy is so low as to be clinically useless (Fig. 1(a)). No increase in study size will change this and no single measure has ever been identified that is clinically useful. Group-level statistical significance is not the same as individual patient clinical significance.
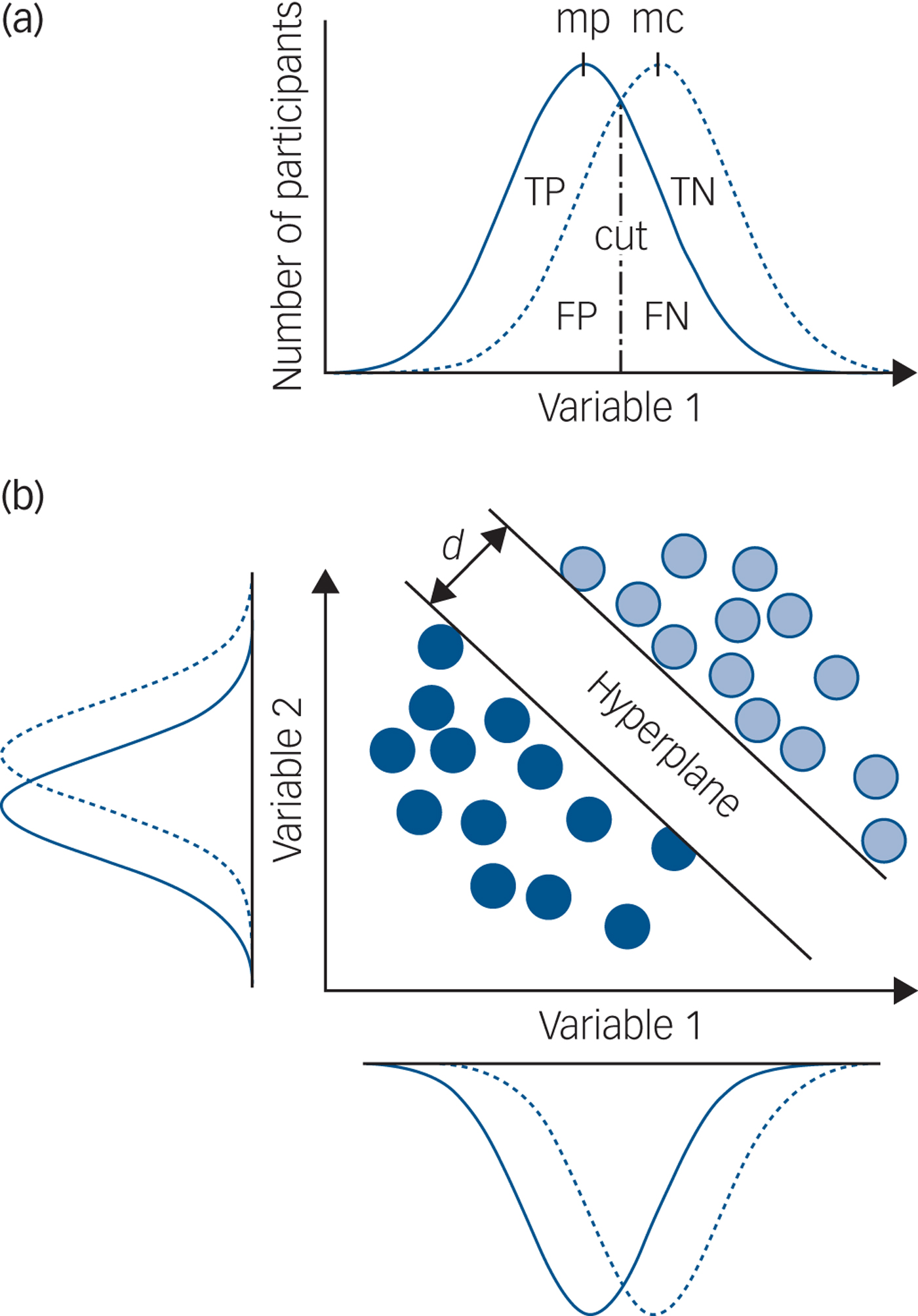
Fig. 1 Group-level significance, univariate and multivariate prediction.
Alternative individual patient pragmatic risk-prediction framework
A multivariate risk-prediction framework may provide a better approach to generate clinically useful predictions about individual patients. The strengths of this approach include not being dependent on first understanding psychiatric nosology, which is controversial, and not requiring formulation of illness ‘mechanisms’ that are yet to be identified. Instead it uses predictors (covariates) to estimate the absolute probability or risk that a certain outcome is present (diagnostic) or will occur within a specific period (prognostic) in an individual with a particular predictor profile, so having an immediate impact on clinical practice.Reference Paulus10 Further strengths of this approach are a clear utilitarian approach, sound statistical basis, a framework for iterative improvement and compatibility with a mechanistic understanding of psychiatric disease.Reference Paulus, Huang, Harle, Redish and Gordon7 This type of approach is already used in other areas of medicine: such as risk calculators for predicting an individual patient's risk of a vascular event or cancer within a given period.
Different computational methods can be used to implement a risk-prediction framework: ‘machine learning’ is a collective term for a set of methods that can be used to train a predictor to make individual patient predictions. At its simplest, a sample of data is split into two parts and one part used to train a predictor with the other part of the data, which has not been used for training, used to test the predictive accuracy, sensitivity and specificity. This is cross-validation, also known as within-study replication. The testing data is equivalent to data from newly presented patients we want to make predictions for. Crucially, even when the distributions of individual variables strongly overlap, it is possible for a machine learning approach to make accurate individual patient predictions (Fig. 1(b)).
Proof of concept studies for individual patient prediction
Prediction methods require quantitative data and these vary in terms of objectivity, practicality and accessibility to routine clinical services. Subjective quantitative data can easily be generated from clinical rating scales, for example the Hamilton Rating Scale for Depression.Reference Hamilton18 These are subjective because patients are asked to match the way they have been feeling to a choice of different statements. There are recent National Health Service (NHS) initiatives to record subjective data for individual patients, such as brief symptom measures acquired as part of the Improving Access to Psychological Therapies (IAPT) in England.Reference Tiffin and Paton19 Rating scales with machine learning have been used to discover subtle patterns of differential treatment response,Reference Koutsouleris, Kahn, Chekroud, Leucht, Falkai and Wobrock20,Reference Chekroud, Zotti, Shehzad, Gueorguieva, Johnson and Trivedi21 although more objective measures might be more reliable predictors and provide better insight into abnormal biology.
Objective data includes items such as age and gender but are of limited use because they contain little information. The most readily available ‘information rich’ source of data is likely a T 1-weighted MRI brain ‘structure’ scan. It is objective because it requires no subjective judgement by a patient and minimal cooperation (lying still). It is practical because it takes ~4 min to acquire and readily available because all NHS radiology departments with an MRI scanner can acquire such data without extra equipment. We provide a few examples of ‘proof of concept’ studies that provide cross-validation estimates for individual patient predictions. If useful for routine clinical practice, further work would be required to develop the technique for an NHS environment.
Example 1: prediction of MDD diagnosis and illness severity
A multicentre study tested if it was possible to make an accurate diagnosis of MDD for individual patients using only T 1-weighted brain ‘structure’ scans.Reference Mwangi, Ebmeier, Matthews and Steele22 A machine learning technique was used and a high diagnostic prediction accuracy of ~90% for individual patients was reported. An independent studyReference Johnston, Steele, Tolomeo, Christmas and Matthews23 reported 85% accuracy of individual patient diagnostic prediction, and using fMRI data, 97% cross-validation diagnostic accuracy.Reference Johnston, Tolomeo, Gradin, Christmas, Matthews and Steele24 Successful prediction of MDD illness severity using T 1 brain ‘structure’ scans has also been reported.Reference Mwangi, Matthews and Steele25
Example 2: prediction of drug-relapse for abstinent methamphetamine users
Accurate prediction of relapse for patients who are currently abstinent but previously drug misusing is not possible using standard clinical measures.Reference Paulus, Huang, Harle, Redish and Gordon7 Fifty-eight methamphetamine dependent patients were recruited from an in-patient treatment programme and participated in a stop-signal task during fMRI. These patients were prospectively followed for a year then reassessed. Using fMRI measures of brain activity in combination with a computational model of behaviour and brain activity, relapse was predicted with accuracy ~78% using cross-validation.Reference Paulus, Huang, Harle, Redish and Gordon7
Example 3: prediction of cognitive–behavioural therapy (CBT) outcome for anxiety disorders
It is difficult to accurately predict outcome of CBT and such treatment requires access to extended therapist time, which is expensive. Before receiving CBT, adults with generalised anxiety disorder or panic disorder took part in an emotional regulation task during fMRI.Reference Ball, Stein, Ramsawh, Campbell-Sills and Paulus26 Standard clinical and demographic variables predicted individual patient clinical outcome with 69% accuracy: fMRI measures predicted individual clinical outcome more accurately at 79%.Reference Ball, Stein, Ramsawh, Campbell-Sills and Paulus26
Example 4: prediction of antidepressant outcome for depression
There is no empirically validated method to determine whether a patient will respond to a specific antidepressant. Using STAR*D ratings and a machine learning approach it was possible to predict individual patient symptomatic remission from a 12-week course of citalopram with 60% accuracy.Reference Chekroud, Zotti, Shehzad, Gueorguieva, Johnson and Trivedi21 The predictive model was validated in an independent sample of patients treated with escitalopram. Further development of predictive techniques to select the best antidepressant for individual patients with MDD would be beneficial. This could be achieved by: (a) including a larger number of questionnaires than STAR*D to identify a better list of questions and (b) use of EEG/MRI measures, which are likely to be useful for predicting antidepressant response.Reference Frodl27
Example 5: prediction of dementia and predictions for non-psychiatric disease
Predicting the risk of dementia for patients presenting with mild cognitive impairment is important clinically as it would (a) facilitate already available preventative interventions for vascular disease (vascular dementia) and (b) for Alzheimer's disease allow recruitment into clinical trials of novel medications for patients who do not have advanced disease. Proof-of-concept studies have been published for some time: for example Zhang & Shen reported 78% accuracy of dementia diagnosis at least 6 months ahead of baseline scanning.Reference Zhang and Shen28 Commercial interest in predictive healthcare computing in non-psychiatric areas is booming: such as Google DeepMind's partnership with the Royal Free London NHS Foundation Trust (acute kidney injury detection) and with University College London hospitals (head/neck cancer and retinal imaging at Moorfields).
Summary and implications
These examples show that using a predictive framework with neuroscience techniques it is possible to make objective clinically useful predictions for individual patients: diagnoses, illness severity, drug abstinence relapse, psychotherapy and medication treatment outcomes. It is important to note that machine learning, like any technique, can be misapplied so care is needed in its use.Reference Arbabshirani, Plis, Sui and Calhoun29–Reference Schnack and Kahn32 Crucially: (a) samples need to be representative of the NHS populations of interest, (b) calculations need to be properly cross-validated and tested against independent samples, (c) different machine learning methods should be compared and (d) careful analyses need to be done to determine whether there are hidden confounders (for example artefacts) driving what the algorithms use to predict.
The ultimate aim of these multivariate approaches is to optimise individual patient care. This means a series of studies are required, starting with relatively inexpensive single studies correctly using cross-validation, then more expensive larger replication studies with independent data, then prospective clinical trials as part of NHS care. There are some initiatives, for example patients with first-episode psychosis in the South London and Maudsley NHS Trust are being assessed using neuroimaging and blood tests as such measures may allow prediction of antipsychotic response.Reference Kempton and McGuire33,Reference McGuire, Sato, Mechelli, Jackowski, Bressan and Zugman34 In addition, these predictive approaches can also be used to aid illness mechanisms research: it is possible to examine the neurobiological differences between those individuals who were successfully predicted to respond to a treatment versus those that were successfully predicted to not respond to a particular treatment. However, to date, no predictive technique for psychiatry is under development in an NHS environment. NHS implementation issues will now be considered.
Implementation in the NHS
It seems inevitable that machine learning based techniques will start to be introduced into non-psychiatric areas of NHS healthcare before long: for example researchers at John Radcliffe Hospital in Oxford have developed a machine learning technique for cardiac scans that greatly outperformed consultants. The route to adoption of new NHS technologies in the UK is well established, first by establishing sufficient evidence for the benefit of the intervention by suitable clinical trials and economic studies of cost feasibility, then assessment of a sufficient level of cost–benefit evidence by the National Institute for Health and Care Excellence (NICE), then recommendation by NICE of adoption by the NHS and finally implementation by the NHS. However, adoption of new techniques is challenging because of the UK government's ‘austerity’ fiscal policy that restricts spending for routine NHS care and other public sector areas and that has been in place for a decade.
The route to NHS implementation of predictive techniques for psychiatric disorders is not fundamentally different; however, there are additional barriers. Quantitative data is required and can be readily generated, but NHS psychiatric services routinely collect very little quantitative data relevant for individual patients. Only patients in old age services presenting with mild cognitive impairment routinely receive an MRI scan as part of assessment. Even for patients presenting with MDD, it is not part of routine practice in general adult psychiatry to quantify illness severity using an established rating scale despite, for example the Hamilton Rating Scale for Depression being available since 1960Reference Hamilton18 although brief symptom measure collection is increasing with IAPT. Instead the UK focus remains qualitative clinical impressions, social interventions and health service reorganisation. Without a change in culture, significant clinical progress seems unlikely.
UK stakeholders, funding and psychiatric research and development (R&D)
UK stakeholders are patients with mental illness and addictions, their families and politically influential organisations representing them. Individual patients are often very supportive of biomedical research into mental illness and addictions, which has allowed recruitment of vast genetics studies and huge neuroimaging studies. However, psychiatry is unique as a medical speciality in having a section of society opposed to a biomedical approach plus staff in community mental health teams do not always accept that mental illness has a biological component. Other than the newly emerged MQ charity, there are no UK mental health charities supportive of biomedical R&D. The Medical Research Council, Wellcome Trust and National Institute for Health Research support biomedical research into psychiatric disorders. However, only approximately 5% of total UK medical funding is spent on mental illnessReference Kirtley35 and a small number of London Universities, Oxford and Cambridge receive ~46% of all UK public and charity funding,36 meaning funding is very limited in most parts of the UK. In contrast, patients with dementia and their charities strongly support biomedical research and funding is far higher.Reference Kirtley35 Therefore, although large numbers of individual patients support biomedical R&D into mental illness and addictions, currently, UK progress in this clinical field is likely to be limited.
Recommendations for training future psychiatrists
(a) It is important for future psychiatrists to understand the value of quantifying clinically relevant aspects of illness, starting by using clinical rating scales and gaining experience in their use. The objective is not to replace qualitative judgement, but to supplement it as is routine in other areas of medicine.
(b) It is crucial for psychiatrists to be able to differentiate statistical significance from clinically meaningful measures useful for individual patient care.
(c) It is helpful to be familiar with RDoC domain constructs and reflect on how psychiatric symptoms for individual patients relate to these: negative and positive valence systems, cognitive systems, social processes and arousal-regulatory systems.Reference Insel, Cuthbert, Garvey, Heinssen, Pine and Quinn16
(d) Learning how to synthesise information across ‘units of analyses’ is important (for example symptoms, behaviour, brain circuits and molecular findings)Reference Insel, Cuthbert, Garvey, Heinssen, Pine and Quinn16 and will help avoid the outdated ‘biology versus psychology’ philosophical dichotomy.Reference Kendler17
(e) To gain entry to medical school, all consultant psychiatrists had the necessary qualifications to study undergraduate mathematics. The potential benefit of quantitative methods for patients should be introduced early in undergraduate medical training.
Conclusions
Mental illness and substance use disorders are the leading cause of disability worldwide, suicide is a leading cause of death in young adults, and severe and enduring mental illness is associated with a reduction in lifespan of about a decade. Despite this, clinical practice in psychiatry has not changed fundamentally in over half a century. There is good evidence that clinically useful individual patient predictions of diagnosis, clinical outcome and treatment response, can be made using neuroscience techniques. This predictive approach does not depend on understanding psychiatric nosology or illness mechanisms.
Given the remarkable disability and mortality associated with mental illness and addictions, it is crucial to invest more in UK biomedical research and clinical practice implementation. However, without organised stakeholder support influencing politicians and funding leaders, not much is likely to change. Currently, the area of UK psychiatry that seems most likely to develop and implement these new clinical techniques is old age psychiatry. We hope this will expand to include mental illness and addictions when the potential benefits become better known.






eLetters
No eLetters have been published for this article.