Despite all controversies in molecular cause and cellular origin of cancer, there is a relative agreement that some physiological and pathological states including ageing, obesity and type 2 diabetes are associated with increased risk of many types of cancer( Reference Berstein1 ). Based on epidemiological studies, obesity correlates with the increase in incidence of several cancer types including the colon, breast, endometrium, oesophagus, pancreas and liver( Reference Basen-Engquist and Chang2 ). Among them, colorectal cancer (CRC) is the third most common cancer in men and women( Reference Siegel, Miller and Jemal3 ). Although some mechanisms such as hyperinsulinaemia and high level of insulin-like growth factor-1 are proposed to link adiposity and cancer( Reference Cohen and LeRoith4 ), obesity-induced chronic inflammation has been further mentioned. The high prevalence of cancer in inflammatory diseases including chronic pancreatitis, ulcerative colitis and liver infectious diseases further confirm the probable involvement of inflammation in obesity-induced tumorigenesis( Reference Deng, Lyon and Bergin5 ).
It was believed that conversion of stem cells into specific type of differentiated cells is a unidirectional way. However, Yamanaka’s pioneering study indicated that artificial expression of few stemness transcription factors (TF), including SOX-2, OCT-4, KLF-4, NANOG, LIN-28A and c-Myc in differentiated somatic cells, is sufficient to reprogram them into pluripotent stem cells( Reference Okita, Ichisaka and Yamanaka6 ).
Although, based on the prevalent paradigm, intestinal stem cells are the main origin of CRC stem cells( Reference Sell7 ), it has been accepted that differentiated epithelium can also acquire precancerous properties (reviewed in Huels & Sansom( Reference Huels and Sansom8 )). Intestinal stem cells are located in mucosal crypt base and undergo self-renewal and differentiation to provide all intestinal cell population. Early observation of physicians indicated that adenomatous polyps, as a preneoplastic lesion, appear at the top of colonic crypts( Reference Cole and McKalen9 ). Furthermore, differentiated intestinal epithelium transforms into neoplastic state in top-down manner during inflammatory bowel disease( Reference Shih, Wang and Traverso10 ). Differentiated cells need to re-enter cell cycle, overcoming tumour suppressor barriers and expressing stemness genes to generate stem-like properties and transform into neoplastic cells( Reference Soufi and Dalton11 ).
TF control cell identity and final fate through transcriptional network of specific cell lineage( Reference Yamamizu, Piao and Sharov Alexei12 ). It has been proposed that NF-κB transcription factor, as main regulator of inflammation, provides mechanistic link between chronic inflammatory conditions and tumorigenesis. Signals that induce inhibitor of NF-κB (IκB) phosphorylation and subsequent degradation, such as proinflamatory cytokines and DNA damage, cause nuclear translocation of NF-κB dimer such as p65/p52( Reference Luo, Kamata and Karin13 ). Nuclear NF-κB promotes the expression of a subset of genes including those involved in inflammation process. Recently, it has been shown that NF-κB is the main culprit in inflammation-associated epithelial-to-mesenchymal transition and stemness( Reference Zhou, Liu and Tang14 ).
Epigenetic landscape of somatic cells is dramatically altered during dedifferentiation. Impairment in epigenetic alterations can disturb cellular dedifferentiation and reprogramming( Reference Papp and Plath15 ). Master stemness factors usually are suppressed in adult somatic tissues( Reference De, Jeong and Leem16 ). It has been previously revealed that inflammatory stimuli remarkably induce alteration in DNA methylation( Reference Ligthart, Marzi and Aslibekyan17 ). We assume that obesity-associated inflammation and NF-κB activation might overcome epigenetic barriers of stemness genes’ expression to reproduce stem-like properties in differentiated epithelial cells. Methylation of cytosine–phosphate diester–guanine (CpG) on transcriptional cis-regulatory elements, namely promoters and enhancers, often suppresses transcription (reviewed in Han( Reference Han18 )). DNA methyltransferases (DNMT) are responsible for DNA methylation, and ten-eleven translocation (TET) enzymes are part of a DNA demethylation mechanism( Reference Jin and Robertson19 , Reference Wu and Zhang20 ).
In this study, we proposed that NF-κB transcription factor might epigenetically control expression of genes involved in dedifferentiation. To this purpose, we analysed methylation of CpG within gene promoters of POU5F1, NANOG and MYC nearby NF-κB binding sites in obese human colonic tissues. In parallel, promoter methylation of CDKN2A was analysed to show DNA methylation status of a tumour suppressor in response to obesity. Moreover, HT-29 cells treated by TNF-α were exploited to show the role of NF-κB in epigenetic alteration of stemness factors. Although there is evidence for the role of NF-κB in dedifferentiation and reprogramming, its contribution in obesity-induced hypomethylation of stemness genes has not been addressed. Here, we provide evidence regarding the probable obesity-related NF-κB activation on hypomethylation of NANOG.
Methods
Subjects
In this hospital-based, case–control study, twenty obese men and twenty sex- and age (with a 5-year interval)-matched lean controls were recruited between February 2015 and April 2016 to ambulatory endoscopy clinic of Moddares hospital. Inclusion criteria were as follows: age 20–60 years old, BMI≥30 kg/m2 for obese cases and BMI<25 kg/m2 for lean controls; lack of any acute or chronic illness; and no daily medication. Individuals with a prior history of colonic neoplasia, colitis, polyposis, previous colon resection and CRC or any other cancers were excluded.
In all, 3 d before the colonoscopy, subjects were on a special diet which included low-fibre foods containing no whole grains, nuts, seeds, dried or raw fruits and vegetables. The day before the procedure, no solid food was allowed and subjects were asked to only consume clear liquids such as clear broth, black tea and coffee, or clear juices (apple, white grape), while on the day all were fasted. Standard optical colonoscopy was performed on all participants by the attending gastroenterologist. In all, four rectal pinch biopsies of colon epithelium were obtained from the mid-rectum using jumbo biopsy forceps. The biopsies were immediately snap-frozen in liquid N2, transported to the laboratory and stored at −80°C until further sample processing.
Weight and height were measured using standard protocols, and BMI was calculated by dividing weight in kg by the square of height in metres. Moreover, to calculate waist:hip ratio, waist circumference (WC) and hip circumference were measured using a non-stretchable tape measure. The standard cut-off values for abdominal obesity in adults were WC ≥102 cm in men( Reference Klein, Allison and Heymsfield21 ). Participants were personally interviewed based on a structured questionnaire to obtain information on age, family history of CRC in a first-degree relative, family history of other cancers, education, occupation, smoking habit, chronic or acute disease history, consumption of multivitamin or other nutritional supplement, and list of medications. The ethics committee at the National Nutrition and Food Technology Research Institute of Shahid Beheshti University of Medical Sciences approved the study protocol (code of ethics committee: IR.SBMU.NNFTRI.REC.1395.50). In addition, written informed consents were obtained from all participants before enrolment.
Cell culture and treatment
The HT-29 human colon adenocarcinoma cells were purchased from the National Cell Bank of Pasteur Institute, Iran. The cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Life Technologies) supplemented with 10 % fetal bovine serum (Gibco, Life Technologies), 1 % penicillin/streptomycin (Sigma) and 2 mm glutamine (Sigma). Cells were kept in 37°C, 5 % CO2 in a humidified incubator until reaching approximately 80 % confluency. For TNF treatment experiment, the HT-29 cells were seeded in twelve-well plates (500×105 cells/well). After 24 h, medium was renewed, and the cells were treated with human recombinant TNF (Invitrogen) at two different concentrations of 10 and 25 ng/ml and incubated for 48 and 72 h.
DNA extraction and sodium bisulphite modification
Total genomic DNA was extracted from the cultured cells and tissue samples using EZ-10 spin column animal DNA mini-preps kit (Bio Basic Inc.) according to the manufacturer’s protocol. In the next step, the total genomic DNA samples were modified by sodium bisulphite conversion using an EZ DNA methylation-gold kit (Zymo Research). The purpose was to convert unmethylated cytosines to uracils while leaving methylated cytosines unmodified. The modified DNA samples were stored immediately at −80°C.
Primer design
High-resolution melting (HRM) primer pairs were designed for specific CpG islands in promoter sequences of each gene according to the HRM primer design guidelines (Table 1). In addition, we aimed to design primers for a given DNA sequence based on their proximity to NF-κB binding site. NF-κB proteins act as transcriptional activators or repressors by binding to the consensus DNA sequence (5-GGGRNYYYCC-3) known as the kB site. We analysed the putative NF-κB binding sites within the promoter region of human POU5F1, NANOG and p-16 promoter using Softberry (http://www.softberry.com/berry.phtml?topic=nsite&group=programs&subgroup=promoter) and MatInspector (http://www.genomatix.de) programs.
Table 1 Methylation-sensitive high-resolution melting primers and amplicon information
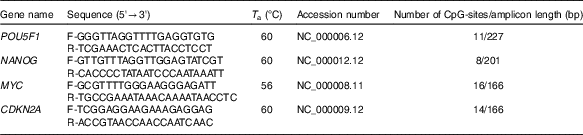
T a, appropriate annealing temperature; CpG, cytosine–phosphate diester–guanine.
Methylation-sensitive high-resolution melting
HRM was used to assess the methylation status of the NANOG, POU5F1, MYC and CDKN2A genes. PCR was carried out in a 20 µl total volume containing 13 µl of double-distilled water, 4 µl of 5× hot FIREPOL Eva Green HRM mix-Rox kit (Solis BioDyne), primers at final concentration of 0·3 pmol and 1 µl of bisulphite modified template. Each reaction was performed in duplicate. HRM was performed using the following protocol: (1) PCR amplification protocol including denaturation for 15 min at 95°C for one cycle, denaturation for 15 s at 95°C, appropriate annealing temperature for each primer set (Table 1) for 15 s and extension for 20 s at 72°C for forty-five cycles; followed by (2) HRM protocol including 95°C for 1 min, 40°C for 1 min, 74°C for 5 s and continuous acquisition to 90°C at twenty-five acquisitions per 1°C (step one plus; Applied Biosystems). Unconverted DNA (not treated with the bisulphite reagent) served as a negative control. Human methylated and unmethylated DNA sets from Zymo Research were used as 100 % methylated and 0 % unmethylated controls, respectively. The percentages of methylation of 0, 25, 50, 75 and 100 % were used to draw the standard curve for the POU5F1, NANOG and CDKN2A genes, Whereas the standards of 0, 5, 10, 25, 50, 75 and 100 % were used for the MYC. Melting curves were normalised relative to two normalisation regions before and after major decrease in fluorescence. This indicated the melting region of the PCR product using the HRM version 2.2 software (ThermoFisher Scientific).
Western blotting analysis
Frozen colon tissue samples were homogenised in radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology), incubated for 30 min on ice and centrifuged for 20 min at 15 000 g. Protein assay was performed using the Bradford method. Equal amount of proteins (50 µg) were subjected to SDS-PAGE and then electroblotted onto a polyvinylidene difluoride membrane. The membranes were blocked with 5 % bovine serum albumin for 1 h at room temperature and incubated with a primary mouse pIκB antibody (B9, Santa Cruz Biotechnology, Inc.) at 4°C for overnight, followed by incubation with a secondary antibody (Santa Cruz Biotechnology Inc.). Subsequently, the signal was detected by chemiluminescence (Amersham ECL kit; GE Healthcare, Life Sciences). In addition, equal loading and constant protein expression of β-actin were determined and served as the normalising control.
Statistical analysis
Statistical tests were performed using SPSS software (version 16.0), and a two-sided P-value ≤0·05 was considered significant. χ 2 Tests or independent-samples t tests were applied for categorical and continuous variables, respectively. Where the distribution of the continuous variables was not normal, the Mann–Whitney U test was used. To assess the relation between obesity (BMI>30 kg/m2) and percentage methylation of the POU5F1, NANOG and MYC logistic regression models were applied for estimating OR with 95 % CI adjusting for age and smoking. Gene methylation was assessed as medians based on the distribution among the controls. We also assessed the relation between methylation status and WC using regression analysis.
Results
Characteristics of cases and controls
Demographic and lifestyle characteristics of participants are presented in Table 2. By design, age was similar in both groups (50·8 v. 52·5 years in controls and cases, respectively). However, the cases were more likely to have a history of former smoking (P≤0·05). No significant differences were observed in terms of cancer and particularly colon cancer family history, non-steroidal anti-inflammatory drug use, marital status and educational level between cases and matched controls.
Table 2 Selected baseline characteristics of cases (twenty) and controls (twenty), Iranian case–control study, 2016–2017 (Mean values and standard deviations; medians and interquartile ranges (IQR); numbers and percentages)
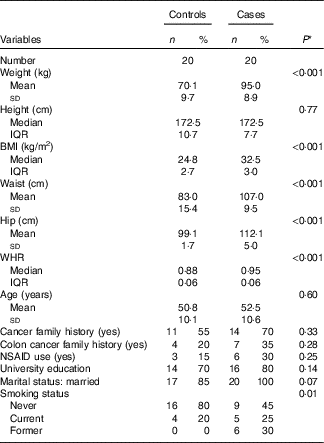
WHR, waist:hip ratio; NSAID, non-steroidal anti-inflammatory drugs.
* Student’s t test or Mann–Whitney test was used for continuous variables with normal and non-normal distribution, respectively, χ 2 test was used for categorical variables.
Phosphorylation of inhibitor of NF-κB in colon epithelial tissue
Due to the small size of obtained colon tissues and the importance of DNA methylation analysis of stemness genes, only phosphorylation of IκB could be analysed as the indicator of subsequent nuclear translocation and activation of NF-κB. As shown in Fig. 1, three distinct immunoblotting analyses on the lysate of the colon tissue samples revealed that IκB phosphorylation was remarkably higher in obese samples in comparison with normal-weight controls. Higher level of IκB phosphorylation was observed across all cases.
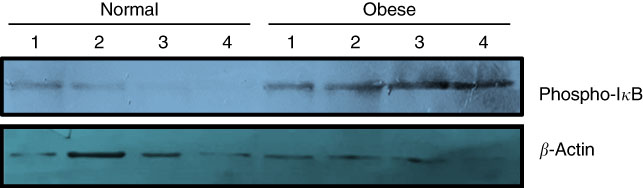
Fig. 1 Phosphorylation of inhibitor of NF-κB (IκB) increases in the colon tissue samples of obese subjects. Frozen colon sample tissues were lysed by radioimmunoprecipitation assay buffer and an equal amount of protein was subjected to Western blot using primary antibodies against total phospho-IκB (Ser 32). Antiactin antibody was used as the loading control. Results are representative of three independent experiments.
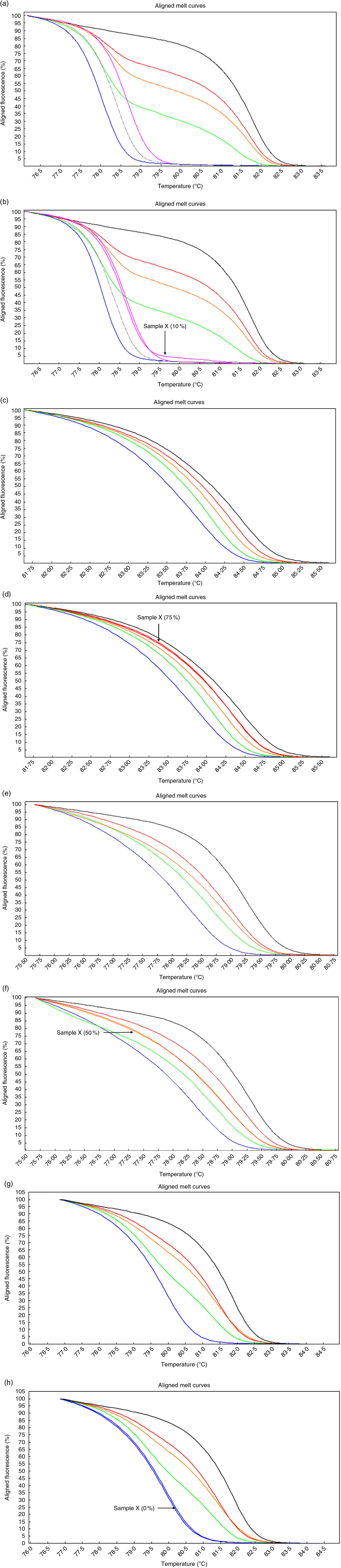
Fig. 2 High-resolution melting aligned melt curves for the MYC, POU5F1, NANOG and CDKN2A methylation. Control curves (a) and one sample curves (b) for the MYC gene. Control curves (c) and one sample curves (d) for the POU5F1 gene. Control curves (e) and one sample curves (f) for the NANOG gene. Control curves (g) and one sample curves (h) for the CDKN2A gene. Standards: 100 %, black line; 75 %, red line; 50 %, orange line; 25 %, green line; 10 %, pink line; 5 %, gray line and 0 %, blue line.
Hypomethylation of NANOG gene in colon epithelial tissue
Based on software analysis, we found that POU5F1, NANOG and MYC promoter regions include a NF-κB binding site (POU5F1: +247/257, NANOG: −485/−475, MYC: +288/298s from transcription start site) whereas CDKN2A lacks NF-κB binding site in its promoter. Preliminary results indicated that methylation of the CDKN2A promoter was rare: 0 % in both the cases and controls. The median methylation levels of the POU5F1, NANOG and MYC in the controls were 50 % interquartile range (IQR: 25), 62·5 % (IQR: 25) and 5 % (IQR: 5), respectively, whereas the levels were 50 % (IQR: 25), 50 % (IQR: 25) and 10 % (IQR: 5) in the obese cases, respectively. Statistical analysis confirmed that the methylation (%) level of the NANOG was significantly different (P<0·05; Mann–Whitney U test) between obese cases and lean controls.
The OR from binary logistic regression models of the association between obesity and gene methylation by median categories of the control groups are presented in Table 3. We did not find any association between obesity and POU5F1 and MYC promoter methylation. However, obesity was more likely to be associated with hypomethylation of NANOG in both crude and adjusted models (crude OR=0·25, 95 % CI 0·06, 1·00; adjusted OR=0·18, 95 % CI 0·04, 0·86). Following categorising the subjects into two groups according to WC (≥102cm as an indicator of central obesity), we found an inverse association between WC and NANOG methylation but only after adjusting for age and smoking (adjusted OR=0·08, 95 % CI 0·008, 0·90).
Table 3 Association of obesity (BMI ≥30 kg/m2), central obesity and genes methylation by median categories of control group in a case–control study(Odds ratios and 95% confidence intervals)
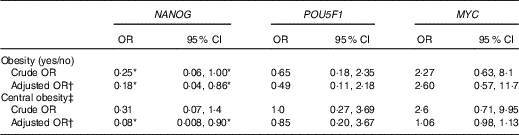
* The association is statistically significant (P≤0·05).
† Adjusted for age and smoking.
‡ Central obesity defined as waist circumference ≥102cm in men and subjects categorised into two groups based on it.
We also examined the association between gene methylation and the characteristics of the study subjects. NANOG methylation status was inversely associated with former smoking (OR=0·07, 95 % CI 0·006, 0·98) after adjusting for age and case–control status (Table 4). We found no other significant associations between genes methylation and other characteristics of the study subjects.
Table 4 Association of NANOG, POU5F1 and MYC methylation status with selected demographic and lifestyle factors of the study subjectsFootnote * (Odds ratios and 95 % confidence intervals)
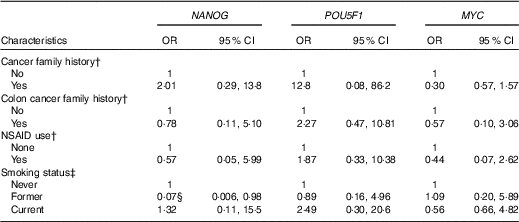
NSAID, non-steroidal anti-inflammatory drugs.
* Genes methylation categorised into two groups based on median categories of control groups.
† Adjusted for age, smoking and case–control status.
‡ Adjusted for age and case–control status.
§ The association is statistically significant (P≤0·05).
Regulation of gene methylation in HT-29 cells by TNF treatment
To mimic the role of NF-κB in hypomethylation of NANOG in the colon epithelial tissue of the obese patients, TNF-α was used as the best known NF-κB stimulator. Methylation-sensitive-HRM assessment of methylation percentage after TNF-α treatment in the HT-29 cell line is shown in Table 5. Two different concentrations of TNF (10, 25 ng/ml) for 48 and 72 h were used. As expected in HT-29 CRC cell line and due to lack of NF-κB binding site in its promoter, CDKN2A was totally hypermethylated (100 %) before and after treatment with TNF-α. The methylation percentages in the control group were stable at all time points. Treatment with 10 ng/ml TNF for 48 h did not affect the methylation status (%) of the POU5F1, NANOG and MYC. However, their methylation (%) was reduced when exposure time increased to 72 h, with the same concentration. Further decline in DNA methylation (%) was observed when 25 ng/ml TNF-α was administered for 48 and 72 h compared with the untreated controls.
Table 5 Methylation status of POU5F1, NANOG and MYC before and after TNF treatments*
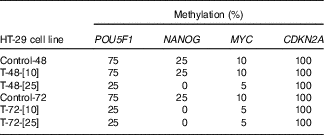
* Control-48 and HT-29 cells were cultured for 48 h in basic medium alone served as the control group. T-48-[Reference Shih, Wang and Traverso10] and HT-29 cells were cultured in basic medium treated with 10 ng/ml TNF for 48 h. T-48-[Reference Peyrin-Biroulet, Chamaillard and Gonzalez25] and HT-29 cells were cultured in basic medium treated with 25 ng/ml TNF for 48 h. Control-72 and HT-29 cells were cultured for 72 h in basic medium alone served as the control. T-72-[Reference Shih, Wang and Traverso10] and HT-29 cells were cultured in basic medium treated with 10 ng/ml TNF for 72 h. T-72-[Reference Peyrin-Biroulet, Chamaillard and Gonzalez25] and HT-29 cells were cultured in basic medium treated with 25 ng/ml TNF for 72 h.
Discussion
Today, it is widely accepted that the cell plasticity allows differentiated cells to revert back to stem cell state. The differentiation states of cells depend on its transcriptional network and related lineage-specific transcription factors. Our study suggested that obesity might attenuate DNA methylation of NANOG in human intestinal mucosa. NANOG plays several roles in promoting tumorigenesis as an oncogene. NANOG is not expressed in normal adult tissues but is highly expressed in some human cancers( Reference Kim, Liu and Qiu22 ). Obesity-induced NANOG hypomethylation may overcome epigenetic barrier of its ectopic expression in colon epithelium. WC is recommended as a stronger risk factor for colon cancer than BMI, as the risk of colon cancer significantly increased with higher levels of central obesity in both men and women( Reference Moore, Bradlee and Singer23 ). Interestingly, our results showed that there is a probable positive correlation between central obesity and hypomethylation of NANOG. NF-κB is the master TF in regulation of inflammation in cells. Recent findings support the idea that obesity and high-fat diet promote intestinal inflammation( Reference Ding and Lund24 ). The association between visceral adiposity and Crohn’s disease has also been reported( Reference Peyrin-Biroulet, Chamaillard and Gonzalez25 ). Accordingly, higher level of phospho-IκB – as an indicator of NF-κB activation – in colonic tissue samples of obese cases, could justify obesity as the cause of inflammation in intestine.
Cutting-edge studies have revealed the role of NF-κB activation in regulation of differentiation. It has been shown that proinflammatory cytokines TNF-α and IL-17 impair osteogenic differentiation of mesenchymal stem cells( Reference Chang, Liu and Lee26 ). Schwitalla et al.( Reference Schwitalla, Fingerle Alexander and Cammareri27 ) showed that intestinal epithelium can be reprogrammed into cancer stem cells through an inflammatory signalling and involvement of Wnt activation. Also, NF-κB has a key role in in vivo dedifferentiation and generation of tumour-initiating cells in intestinal tumorigenesis. Consistently, there is an evidence that NF-κB plays a major role in maintaining the undifferentiated state of human-inducible pluripotent stem cells through up-regulation of NANOG and POU5F1 ( Reference Takase, Yoshikawa and Idei28 ). However, these studies do not provide evidence about the role of NF-κB in transactivation of genes which epigenetically were suppressed in normal tissues.
TET methylcytosine dioxygenases are the main part of active DNA demethylation mechanism( Reference Rasmussen and Helin29 ). TET are not site-specific enzymes to remove methyl moiety from particular promoter; therefore, it might be plausible to assume a molecular guide directing them to specific target genes. Transcription factors seem to be good candidates to do this mission. There are evidences showing some TF regulate DNA methylation through recruitment of DNMT and TET as co-regulators of TF( Reference Brenner, Deplus and Didelot30 ). In addition, differentially methylated regions of DNA were often nearby TF binding sites( Reference Oda, Kumaki and Shigeta31 ). It has been shown that inactivation of repressor element 1-silencing transcription factor (REST) promotes DNA methylation in CpG, which are normally near the REST binding sites. Re-expression of REST induces hypomethylation of these regions. Recently, one study indicated that REST recruits TET3 and subsequently promotes its hydroxylase activity( Reference Perera, Eisen and Wagner32 ). Although the results of the present study did not show direct interaction between NF-κB and TET in demethylation of NANOG or other stemness genes, TNF-α-induced hypomethylation of the NANOG, POU5F1 and MYC genes in the HT-29 cell persuaded us to suggest a role for NF-κB in this process. Further studies are in progress in our laboratory to show the direct interaction between NF-κB and TET.
Hypermethylation of tumour suppressors is a major step in tumorigenesis( Reference Waki, Tamura and Tsuchiya33 ). As mentioned, CDKN2A lacks NF-κB binding sites in its promoter, accordingly TNF-α had no effect on methylation of hypermethylated CDKN2A promoter in HT-29 cells. Considering the fact that our colonic biopsy samples in obese and normal-weight control were non-cancerous, it seems reasonable that in both groups, CDKN2A was completely unmethylated.
TF have pharmacological inhibitors and activators; the suggested role for transcription factors such as NF-κB in DNA methylation regulation provides a novel perspective for developing epigenetic drugs, which may preferentially alter DNA methylation of related genes.
To assess the complex interactions between potential aetiologic factors, molecular alterations and disease evolution, ‘molecular pathology’ and ‘epidemiology’ have recently become integrated, creating the interdisciplinary field of ‘molecular pathological epidemiology’ (MPE). MPE takes a comprehensive approach to incorporate multiple intersecting pathways such as genomics, epigenetics, energetics, inflammation, microbiome, microbiota, immunity and so on into an integrated analysis of aetiologies, evolution and progression of cancers and all other human diseases( Reference Ogino, Chan and Fuchs34 , Reference Ogino, Nowak and Hamada35 ). Our results regarding obesity-driven DNA methylation alterations of NANOG gene in normal colon tissues could be applied in the context of the MPE of cancer epigenetics to identify the possible risk factors contributing to CRC development.
In analysing results, it is important to pay more attention to the level of statistical significance. In the present study, we considered statistical significance at P-value ≤0·05, which was in accordance with most previous studies assessing specific genes( Reference Johnson, Koestler and Cheng36 , Reference Crujeiras, Morcillo and Diaz-Lagares37 ). Recently, it has been suggested that changing the default P-value threshold for statistical significance from 0·05 to 0·005 is needed for improving the reproducibility of scientific research( Reference Benjamin, Berger and Johannesson38 ). However, it seems that reducing P-value level would be more applicable in microarray analysis when multiple tests are conducted on 1000’s of transcripts, in order to reduce chance of false positives( Reference Mudge, Martyniuk and Houlahan39 ). We just assessed the methylation status of four specific genes, and therefore, lower P-value might not be necessary for interpretation our findings.
In conclusion, our findings suggest that obesity and particularly central obesity altered methylation of the NANOG gene in epithelium of the colon. Hypomethylation of this gene occurred in promoter regions close to NF-κB binding sites, suggesting the involvement of this transcription factor in epigenetic regulation. Also, incubation of the HT-29 cells with TNF-α promoted hypomethylation of CpG in the promoter of NANOG, POU5F1 and MYC nearby NF-κB binding sites.
Acknowledgement
The data were originated from the results of an approved doctoral thesis project of National Nutrition and Food Technology Research Institute and Faculty of Nutrition and Food Technology, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Grant sponsor: National Nutrition and Food Technology Research Institute, Tehran, Iran (project no. 635 and contract no. 311-1848).
F. S. performed the main experiments. M. C., K. P., L. T. T., A. M. and R. T. assisted in all the experiments. S. A. H. assisted with project design. H. Z. designed the study and wrote the manuscript.
The authors declare that there are no conflicts of interest.












