1. Introduction
The 9p21 chromosomal region is a well-characterized locus showing association with coronary artery disease (CAD). Several genome wide association studies (GWAS; Helgadottir et al., Reference Helgadottir, Thorleifsson, Manolescu, Gretarsdottir, Blondal, Jonasdottir, Jonasdottir, Sigurdsson, Baker, Palsson, Masson, Gudbjartsson, Magnusson, Andersen, Levey, Backman, Matthiasdottir, Jonsdottir, Palsson, Einarsdottir, Gunnarsdottir, Gylfason, Vaccarino, Hooper, Reilly, Granger, Austin, Rader, Shah, Quyyumi, Gulcher, Thorgeirsson, Thorsteinsdottir, Kong and Stefansson2007; McPherson et al., Reference McPherson, Pertsemlidis, Kavaslar, Stewart, Roberts, Cox, Hinds, Pennacchio, Tybjaerg-Hansen, Folsom, Boerwinkle, Hobbs and Cohen2007; Samani et al., Reference Samani, Erdmann, Hall, Hengstenberg, Mangino, Mayer, Dixon, Meitinger, Braund, Wichmann, Barrett, König, Stevens, Szymczak, Tregouet, Iles, Pahlke, Pollard, Lieb, Cambien, Fischer, Ouwehand, Blankenberg, Balmforth, Baessler, Ball, Strom, Braenne, Gieger, Deloukas, Tobin, Ziegler, Thompson and Schunkert2007) and meta-analysis (Palomaki et al., Reference Palomaki, Melillo and Bradley2010; Preuss et al., Reference Preuss, König, Thompson, Erdmann, Absher, Assimes, Blankenberg, Boerwinkle, Chen, Cupples, Hall, Halperin, Hengstenberg, Holm, Laaksonen, Li, März, McPherson, Musunuru, Nelson, Burnett, Epstein, O'Donnell, Quertermous, Rader, Roberts, Schillert, Stefansson, Stewart, Thorleifsson, Voight, Wells, Ziegler, Kathiresan, Reilly, Samani and Schunkert2010) have implicated this region with increased risk of CAD, MI as well as progression of disease. Replication studies with different SNPs in this region in diverse populations including Irish (Meng et al., Reference Meng, Hughes, Patterson, Belton, Kee and McKeown2008), Japanese (Hinohara et al., Reference Hinohara, Nakajima, Takahashi, Hohda, Sasaoka, Nakahara, Chida, Sawabe, Arimura, Sato, Lee, Ban, Yasunami, Park, Izumi and Kimura2008; Hiura et al., Reference Hiura, Fukushima, Yuno, Sawamura, Kokubo, Okamura, Tomoike, Goto, Nonogi, Takahashi and Iwai2008), Koreans (Hinohara et al., Reference Hinohara, Nakajima, Takahashi, Hohda, Sasaoka, Nakahara, Chida, Sawabe, Arimura, Sato, Lee, Ban, Yasunami, Park, Izumi and Kimura2008), Indians (AshokKumar et al., Reference AshokKumar, Emmanuel, Dhandapany, Rani, SaiBabu, Cherian and Thangaraj2011; Bhanushali et al., Reference Bhanushali, Parmar, Contractor, Shah and Das2011), Chinese (Ding et al., Reference Ding, Xu, Wang, Wang, Zhang, Tu, Yan, Wang, Hui, Wang and Wang2009), etc. have corroborated these findings. In addition, SNPs in this locus have also been associated with increased risk of abdominal aortic aneurysm (AAA) (Bown et al., Reference Bown, Braund, Thompson, London, Samani and Sayers2008) and ischaemic stroke (Smith et al., Reference Smith, Melander, Lövkvist, Hedblad, Engström, Nilsson, Carlson, Berglund, Norrving and Lindgren2009).
However, the association of 9p21 SNPs with the extent or severity of CAD has been disputable with some studies stating that 9p21 SNP under study predicts severity of the disease (Ye et al., Reference Ye, Willeit, Kronenberg, Xu and Kiechl2008; Dandona et al., Reference Dandona, Stewart, Chen, Williams, So, O'Brien, Glover, Lemay, Assogba, Vo, Wang, Labinaz, Wells, McPherson and Roberts2010) and another the contrary (Chen et al., Reference Chen, Ballantyne, Gotto and Marian2009). The study by Anderson et al. (Reference Anderson, Horne, Kolek, Muhlestein, Mower, Park, May, Camp and Carlquist2008) also concludes that variants at the 9p21 locus robustly predict angiographic CAD prevalence, but not CAD extent or myocardial infarction. These findings have led to suggestions that the 9p21 locus acts at an early stage (as an ‘initiator’) of coronary atherosclerosis (Horne et al., Reference Horne, Carlquist, Muhlestein, Bair and Anderson2008).
In the Indian context, there have been a few studies that have evaluated genetic variants at this locus. In a study from South India (AshokKumar et al., Reference AshokKumar, Emmanuel, Dhandapany, Rani, SaiBabu, Cherian and Thangaraj2011), GWAS identified SNPs rs2383207 and rs10757278 at the 9p21 locus and nine additional flanking SNPs were evaluated for increased risk of CAD. In the North Indian population study (Kumar et al., Reference Kumar, Yumnam, Basu, Ghosh, Garg, Karthikeyan and Sengupta2010), six SNPs were genotyped in a case-control association study in which three SNPs (rs10116277, rs1333040 and rs2383206) present at the locus 9p21 were significantly associated with CAD. Two other studies have also found increased risk of CAD with the rs10757278 variant (Maitra et al., Reference Maitra, Shanker, Dash, John, Sannappa, Rao, Ramanna and Kakkar2009; Bhanushali et al., Reference Bhanushali, Parmar, Contractor, Shah and Das2011).
However to date, there have been no data on the rs1333049G > C variant regarding its frequency as well as association with CAD risk in the Indian population. The rs1333049 G > C on the 9p21 locus had the strongest association in the Wellcome Trust Case Control consortium (WTCCC, 2007) study and German myocardial infarction (MI) family study (P = 1·80 × 10−14 and 3·40 × 10−6, respectively). This SNP has also been associated with recurrent MI and cardiac death (Buysschaert et al., Reference Buysschaert, Carruthers, Dunbar, Peuteman, Rietzschel, Belmans, Hedley, De Meyer, Budaj, Van de Werf, Lambrechts and Fox2010).
The high propensity of Indians to develop CAD necessitates that we conduct studies on the genetic markers associated with increased risk in this population. Hence, the current study was conducted with the aim of determining the frequency of this SNP as well as association with CAD risk in a select western Indian region (Indo Europeans).
2. Materials and methods
(i) Subjects
The study was performed on a total of 365 unrelated individuals, which consisted of 229 patients with CAD confirmed by coronary angiography: > 50% stenosis in one or more arteries and stable or unstable angina and 151 controls: examined clinically and investigated by electrocardiography and treadmill test to exclude CAD. Informed consent was obtained from all the subjects. The study is in accordance with the Helsinki declaration and was approved by the local ethical committee. A detailed case record form pertaining to information on demographics, medical history and coronary risk factors such as presence of diabetes, hypertension, smoking, lifestyle and current medication was completed for each participant through personal interviews and through perusal of their medical records.
Blood specimens were collected after an overnight fast of 12 h by veni puncture using the vacutainer system from Becton Dickinson (Franklin Lakes, NJ, USA) in the anticoagulant EDTA as well as plain bulb for serum. Serum, EDTA plasma samples were separated by centrifugation and aliquots were preserved at −20°C until analysis.
(ii) Biochemical parameters
The laboratory parameters, namely serum total cholesterol (TC), triglyceride (TG) and high-density lipoprotein cholesterol (HDL-C) levels were determined by routine enzymatic endpoint methods (X Imola; Randox Laboratories Ltd., UK). Low-density lipoprotein-cholesterol (LDL-C) and VLDL cholesterol were calculated according to Friedwald´s formula.
(iii) SNP genotyping
The genomic DNA was isolated from peripheral blood using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). DNA yield and purity was determined by measuring absorbance at 260/280 nm. The rs1333049 genotypes were determined for all study participants using an allele-specific real-time PCR Taqman genotyping assay. The DNA was standardized to 10 ng/ml.
(iv) Statistical analysis
Allele frequency was calculated as the number of occurrences of the test allele in the population divided by the total number of alleles. Any deviation of the genotype frequencies from the Hardy–Weinberg equilibrium (HWE) was assessed by Fischer's exact test. Chi-square (χ) tests were used for comparison of binary variables across groups. The non-parametric Kolmogorov–Smirnov test was used as test of normality for the quantitative variables, those parameters that did not show a normal distribution were log transformed during regression analysis. To determine risk for CAD unadjusted odds ratios were determined by Cochran–Mantel–Haenszel statistics and adjusted odds ratio by multivariate logistic regression. SNPStat online software tool was applied to determine the association of the 9p21 variant rs1333049 with CAD and also to determine the model of association, if any (Sole et al., Reference Sole, Guino, Valls, Iniesta and Moreno2006). Routine statistical analysis were carried out with the SPSS v 15 software (SPSS Inc., Chicago, IL) and GraphPad StatMate 2.0 (GraphPad software Inc) was used to determine the power of the study. Under the significance level of P = 0·05, minor allele frequency between 0·25 and 0·40, assuming population disease prevalence between 5 and 10% and main genetic effect between 1·5 and 1·2, our study design can reach > 85% power when the relative risk (RR) is 1·5 and 50% when it is 1·2.
3. Results
Table 1 displays means and standard deviations for the study subjects for relevant biochemical characteristics as well as risk factors. Statistically significant differences were seen in the smoking status (P < 0·01), gender (P < 0·0001), presence of family history (P < 0·0001), diabetes (P < 0·0001) as well as hypertension (P < 0·0001) in cases versus controls. Genotypic and allelic frequencies of the rs1333049 SNP are shown in Table 2. No significant departure from the HWE was seen. A higher frequency of the rs1333049 C allele was seen in cases as compared with the controls (0·60 versus 0·49), which was significant (OR = 1·564, 95%CI = 1·154–2·119, P = 0·003) in the univariate analysis. The univariate analysis also indicated a much higher OR on comparison of CC versus GG genotype (OR = 2·814, 95%CI = 1·426–5·578, P value = 0·001). In the logistic regression analysis, female gender was associated with decreased CAD risk (OR = 0·241, 95%CI = 0·092–0·062, P = 0·004) even after accounting for the differences in their ratio in cases and controls, whereas rs1333049 with increased CAD risk (OR = 2·460, 95%CI = 1·139–5·314, P = 0·022) as seen in Table 3. To determine if this variant is associated with age of onset of the disease chi-square analysis was done (Table 4). A higher frequency of the risk allele was seen in the younger age group with 40% of individuals in age group 30–55 years having the CC genotype as compared with 29 and 24·5% in age group 56–65 years and > 65 years, respectively. These findings were significant but only when the CC genotype was compared with the GG genotype. This may be indicative of a recessive model of inheritance, where lack of the G allele may be responsible for the disease risk per se. The association of rs1333049 (C allele) with the end-point of non-fatal MI was also determined (Table 5). Although a higher frequency of the CC genotype was seen in MI patients, it was not statistically significant (allelic OR = 1·361, 95%CI = 0·954–1·942, P = 0·084). However in agreement with the previous table (Table 2), the combined OR of MI+CAD versus controls was highly significant (OR = 1·540, 95%CI = 1·333–2·094, P = 0·005). The rs1333049 SNP was also in strong linkage disequilibrium with the rs10757578 SNP (previously studied) (Bhanushali et al., Reference Bhanushali, Parmar, Contractor, Shah and Das2011) with D' statistic, 0·794; r statistic, 0·726 and P < 0·01. The haplotype frequencies are indicated in Table 6. In LD block (rs1333049, rs10757278), the two most frequent haplotypes accounted for > 85% of the chromosomes (Hap 1, CG 43 and 56% in controls and cases respectively and Hap 2, GA 42 and 31% in controls and cases, respectively).
Table 1. Clinical and demographic details of the study population
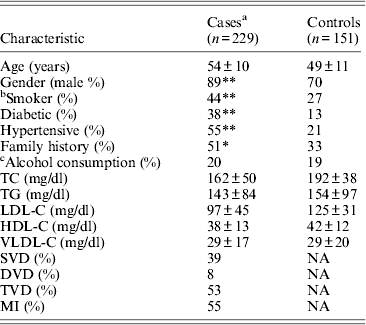
Mean ± SD for continuous variables. SD, standard deviation; NA, not applicable; TC, total cholesterol; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; VLDL-C, very-low-density lipoprotein cholesterol; APOA1, apolipoprotein A1; APOB, apolipoprotein B; TG, triglycerides; SVD, single-vessel disease; DVD, double-vessel disease; TVD, triple-vessel disease; MVD, multiple-vessel disease; MI, myocardial infarction.
*P < 0·001; **P < 0·0001.
a Includes individuals on lipid lowering medications.
b Smokers includes ex-smokers for more than 5 years.
c Alcohol consumption indicates individuals with > 5–7 pegs/week.
Table 2. Genotypic and allelic frequency of 9p21 (rs1333049) in cases and controls

SNP, single-nucleotide polymorphism OR aUnadjusted Odds ratio derived using Cochran–Mantel–Haenszel statistics. 95% CI, 95% Confidence Interval. Statistically significant at *P < 0·05, **P < 0·01.
Table 3. Multivariate logistic regression analysis of association of rs1333049 with CAD risk
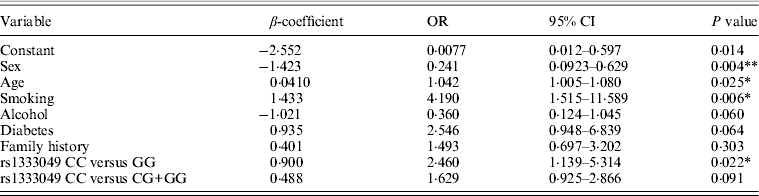
*P < 0·05, **P < 0·005.
Table 4. Association of rs1333049 with age of onset
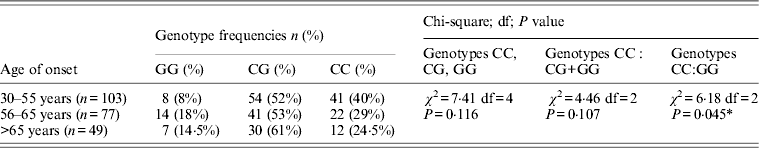
χ 2, Chi-square; df, degree of freedom; *Statistically significant at P < 0·05.
Table 5. Genotype frequency and association of rs1333049G > C with MI as endpoint
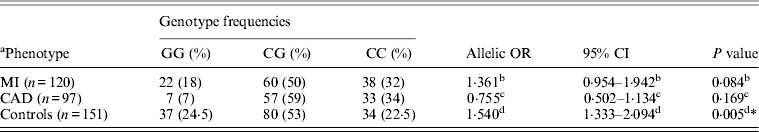
* Statistically significant at P < 0·01.
a Data on 217 individuals (cases), for 12 patients with ischaemic heart disease it was not clear if the endpoint was MI.
b OR, 95% CI, P value indicate OR, 95% CI and P value for MI versus controls.
c OR, 95% CI, P value indicate OR, 95% CI and P value for MI versus CAD.
d OR, 95% CI, P value indicate OR, 95% CI and P value for MI+CAD versus controls.
Table 6. Haplotype frequencies of rs1333049G > C and rs10727578A > G
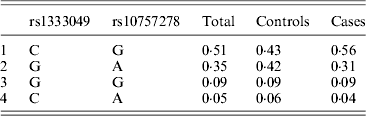
4. Discussion
The magnitude of CAD represents a major socioeconomic impact especially in the developing nations of South-Asia (Yusuf et al., Reference Yusuf, Reddy, Ounpuu and Anand2001; Gaziano et al., Reference Gaziano, Bitton, Anand, Abrahams-Gessel and Murphy2010). It is well known that CAD arises from a combination of environmental and genetic factors (Yusuf et al., Reference Yusuf, Hawken, Ounpuu, Dans, Avezum, Lanas, McQueen, Budaj, Pais, Varigos and Lisheng2004; Damani & Topol, Reference Damani and Topol2007). To date several GWA studies have highlighted the importance of the 9p21 chromosomal region with CAD/MI risk (Helgadottir et al., Reference Helgadottir, Thorleifsson, Manolescu, Gretarsdottir, Blondal, Jonasdottir, Jonasdottir, Sigurdsson, Baker, Palsson, Masson, Gudbjartsson, Magnusson, Andersen, Levey, Backman, Matthiasdottir, Jonsdottir, Palsson, Einarsdottir, Gunnarsdottir, Gylfason, Vaccarino, Hooper, Reilly, Granger, Austin, Rader, Shah, Quyyumi, Gulcher, Thorgeirsson, Thorsteinsdottir, Kong and Stefansson2007; McPherson et al., Reference McPherson, Pertsemlidis, Kavaslar, Stewart, Roberts, Cox, Hinds, Pennacchio, Tybjaerg-Hansen, Folsom, Boerwinkle, Hobbs and Cohen2007; Samani et al., Reference Samani, Erdmann, Hall, Hengstenberg, Mangino, Mayer, Dixon, Meitinger, Braund, Wichmann, Barrett, König, Stevens, Szymczak, Tregouet, Iles, Pahlke, Pollard, Lieb, Cambien, Fischer, Ouwehand, Blankenberg, Balmforth, Baessler, Ball, Strom, Braenne, Gieger, Deloukas, Tobin, Ziegler, Thompson and Schunkert2007; O'Donnell et al., Reference O'Donnell, Kavousi, Smith, Kardia, Feitosa, Hwang, Sun, Province, Aspelund, Dehghan, Hoffmann, Bielak, Zhang, Eiriksdottir, van Duijn, Fox, de Andrade, Kraja, Sigurdsson, Elias-Smale, Murabito, Launer, van der Lugt, Kathiresan, Krestin, Herrington, Howard, Liu, Post, Mitchell, O'Connell, Shen, Shuldiner, Altshuler, Elosua, Salomaa, Schwartz, Siscovick, Voight, Bis, Glazer, Psaty, Boerwinkle, Heiss, Blankenberg, Zeller, Wild, Schnabel, Schillert, Ziegler, Münzel, White, Rotter, Nalls, Oudkerk, Johnson, Newman, Uitterlinden, Massaro, Cunningham, Harris, Hofman, Peyser, Borecki, Cupples, Gudnason and Witteman2011). However, it is crucial to replicate these findings in independent studies as well as to extend and verify its validity in different ethnic populations. The rs1333049 was identified in WTCCC GWA study as the strongest SNP associated with CAD risk (WTCCC, 2007). Although other variants at the 9p21 locus have been studied in Indians (Kumar et al., Reference Kumar, Yumnam, Basu, Ghosh, Garg, Karthikeyan and Sengupta2010; AshokKumar et al., Reference AshokKumar, Emmanuel, Dhandapany, Rani, SaiBabu, Cherian and Thangaraj2011; Bhanushali et al., Reference Bhanushali, Parmar, Contractor, Shah and Das2011; Maitra et al., Reference Maitra, Shanker, Dash, John, Sannappa, Rao, Ramanna and Kakkar2009), no information is available to date on this variant. The results of our study determine for the first time the frequency of this SNP in the select population from Western India as well as confirm the association with CAD risk. The study also sheds light on the LD of rs1333049 with rs10757278 which was found to be associated in another GWA study (Helgadottir et al., Reference Helgadottir, Thorleifsson, Manolescu, Gretarsdottir, Blondal, Jonasdottir, Jonasdottir, Sigurdsson, Baker, Palsson, Masson, Gudbjartsson, Magnusson, Andersen, Levey, Backman, Matthiasdottir, Jonsdottir, Palsson, Einarsdottir, Gunnarsdottir, Gylfason, Vaccarino, Hooper, Reilly, Granger, Austin, Rader, Shah, Quyyumi, Gulcher, Thorgeirsson, Thorsteinsdottir, Kong and Stefansson2007).
In the present study, the C allele of the rs1333049 SNP has a frequency of 0·49 in controls which is similar to that seen in Europeans (0·49), Han Chinese (0·48) and Japanese (0·51) but significantly different to the Yoruban population (0·17) and Kenyan population (0·24) (HapMap data).
Although the rs1333049 was seen to be in strong LD with the rs10757278 (previously studied), D′ = 0·796 however the haplotype structure was not in near complete LD as seen in another study where the D′ was 0·99 (Chen et al., Reference Chen, Ballantyne, Gotto and Marian2009). The haplotype structure in our population is also at variance with the HapMap CEU population where the rs10757278 and this SNP are practically equivalent with linkage r 2 = 1 (HapMap data), unlike in our population where the r statistic is 0·726.
The results of our study confirm that the rs1333049 is significantly associated with CAD risk in the Indian population, which is in agreement with findings in other ethnic populations. In the present study, the OR was 1·564, 95% CI = 1·154–2·119, P = 0·003 in the allele count model, which is higher than that seen in the Japanese (OR = 1·30, 95%CI = 1·13–1·49, P = 0·00027) or Koreans (OR = 1·09, 95%CI = 1·02–1·38, P = 0·025) in the same model (Hinohara et al., Reference Hinohara, Nakajima, Takahashi, Hohda, Sasaoka, Nakahara, Chida, Sawabe, Arimura, Sato, Lee, Ban, Yasunami, Park, Izumi and Kimura2008). Furthermore, comparison of the CC genotype versus the GG genotype indicated a much higher OR = 2·814, 95%CI = 1·426–5·578, P = 0·001. In the discovery GWA study the OR for the WTCCC and the German data were 1·37 and 1·33 for the risk allele (95% CI = 1·26–1·48, P = 1·80 × 10−14 and 95% CI = 1·18–1·51, P = 6·80 × 10−6, respectively).
When the cases were further differentiated based on age a much higher frequency of the CC genotype was seen in the premature CAD group (30–55 years). Similar findings were seen in the Irish population (Meng et al., Reference Meng, Hughes, Patterson, Belton, Kee and McKeown2008) where three SNPs at the 9p21 locus, which included the rs1333039 (P = 3·08 × 10−7) were strongly associated with early onset CAD. The rs1333049 polymorphism was also associated with an earlier age of disease onset in two coronary disease cohorts (Ellis et al., Reference Ellis, Pilbrow, Frampton, Doughty, Whalley, Ellis, Palmer, Skelton, Yandle, Palmer, Troughton, Richards and Cameron2010). The coronary disease cohort study (CDCS) patients homozygous for the high-risk C allele had an age of onset 2–5 years earlier for coronary disease (P = 0·005), angina (P = 0·025), MI (P = 0·022) and percutaneous transluminal coronary angioplasty (P = 0·009). The post myocardial infarction (PMI) participants with the CC genotype were 3 years younger on admission (P = 0·009). The homozygous CC genotype of rs1333049 was also seen to confer a magnified risk of early-onset and severe CAD in diabetic patients (Wang et al., Reference Wang, Peng, Lu, Zhang, Zhang, Wang, Chen and Shen2011). Statistically significant association between 9p21 SNPs and heart disease that varied by age at disease onset was also seen in a meta-analysis (Palomaki et al., Reference Palomaki, Melillo and Bradley2010) although the magnitude of association was small.
Although the association of the 9p21 region with CAD risk is well established, the association with clinical outcomes still remains unclear. The Global Registry of Acute Coronary Events Genetics (GRACE) study found the rs1333049 at-risk C allele to be significantly and independently associated with recurrent MI. Moreover, inclusion of the rs1333049 into the GRACE risk score was also seen to improve classification for recurrent MI or cardiac death (Buysschaert et al., Reference Buysschaert, Carruthers, Dunbar, Peuteman, Rietzschel, Belmans, Hedley, De Meyer, Budaj, Van de Werf, Lambrechts and Fox2010). Similarly strong association of 6 SNPs at the 9p21 locus (rs1333049 included) with MI was also seen particularly in patients with a positive family history (Scheffold et al., Reference Scheffold, Kullmann, Huge, Binner, Ochs, Schöls, Thale, Motz, Hegge, Stellbrink, Dorsel, Gülker, Heuer, Dinh and Stoll2011). These findings were in agreement with a GWA study, where the rs1333049 was strongly associated (P = 7·58 × 10−19) with coronary artery calcification (CAC) and MI (O'Donnell, 2011). The meta-analysis by Preuss et al. (Reference Preuss, König, Thompson, Erdmann, Absher, Assimes, Blankenberg, Boerwinkle, Chen, Cupples, Hall, Halperin, Hengstenberg, Holm, Laaksonen, Li, März, McPherson, Musunuru, Nelson, Burnett, Epstein, O'Donnell, Quertermous, Rader, Roberts, Schillert, Stefansson, Stewart, Thorleifsson, Voight, Wells, Ziegler, Kathiresan, Reilly, Samani and Schunkert2010) indicated that the rs1333049 conferred a 29% increase in risk for MI per copy (P = 2 × 10−20) (Preuss et al., Reference Preuss, König, Thompson, Erdmann, Absher, Assimes, Blankenberg, Boerwinkle, Chen, Cupples, Hall, Halperin, Hengstenberg, Holm, Laaksonen, Li, März, McPherson, Musunuru, Nelson, Burnett, Epstein, O'Donnell, Quertermous, Rader, Roberts, Schillert, Stefansson, Stewart, Thorleifsson, Voight, Wells, Ziegler, Kathiresan, Reilly, Samani and Schunkert2010).
In contrast the association of 9p21 SNPs (rs2383206, rs2383207, rs10757274 and rs10757278) with occurrence of nonfatal MI or death could not be established by Horne and colleagues (Horne et al., Reference Horne, Carlquist, Muhlestein, Bair and Anderson2008). Similarly in another study though the rs1333049 (C) was associated with greater atherosclerotic plaque, it did not associate with MI when disease severity was accounted for (Dandona et al., Reference Dandona, Stewart, Chen, Williams, So, O'Brien, Glover, Lemay, Assogba, Vo, Wang, Labinaz, Wells, McPherson and Roberts2010). Variable results have also been seen with respect to disease progression with one study indicating the sequence variation in this locus to influence atherosclerosis development and progression (Ye et al., Reference Ye, Willeit, Kronenberg, Xu and Kiechl2008) and another indicating no effect either on disease severity or progression (Chen et al., Reference Chen, Ballantyne, Gotto and Marian2009). In the present study, also no significant association of the risk allele with MI was seen. The conflicting results seen to date in phenotypic studies indicate that still more needs to be done to understand the direct as well as indirect mechanisms underlying the effects of this locus.
In conclusion, the rs1333049 SNP at the 9p21 locus is not only significantly associated with CAD risk, which is in concordance with results across several other ethnic groups, but also with age of onset in the Western Indian population. However, there are differences in the haplotype structure of this SNP with the neighbouring rs10757278 SNP. These differences emphasize the importance of genotyping all risk variants at this locus, as it could underlie the differences in risk susceptibility to CAD, which varies across populations. More importantly the high propensity of Indians to develop CAD emphasizes the need for genetic studies in this population. This can also have implications for prevention as adding the 9p21 allele to traditional risk factors was seen to modestly improve CAD risk prediction in the intermediate categories in the population studied (Brautbar et al., Reference Brautbar, Ballantyne, Lawson, Nambi, Chambless, Folsom, Willerson and Boerwinkle2009).
Thus although the study has limitations, that of sample size and consequently the statistical power to examine multiple interactions, these findings provide for the first time data on frequency and association of the rs1333049 variant with CAD and larger studies are warranted in this high-risk population.
Financial Support
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Statement of Interest
None.










