NF-κB is one of the most important signalling pathways linked to the loss of skeletal muscle mass in normal physiological and pathophysiological conditions(Reference Li, Malhotra and Kumar1). NF-κB is constitutively expressed and exists in the cytosol as part of a heterotrimeric complex(Reference Schreiber, Nikolaus and Hampe2). This complex typically comprises the DNA-binding proteins p50 and p65 and the inhibitory protein inhibitor of κBα (IκBα). Activation of NF-κB requires phosphorylation of IκBα, followed by ubiquitin conjugation and proteolysis of IκBα by the 26S proteasome(Reference Jové, Planavila and Laguna3, Reference Chung, Brown and Provo4). The activated NF-κB dimer is then translocated to the cell nucleus, where it regulates the expression of specific genes, such as the gene muscle RING finger 1 (MuRF1)(Reference Cai, Frantz and Tawa5).
MuRF1 is found in the nuclei of muscle cells and has been demonstrated to have ubiquitin-ligase activity, which depends on the presence of the RING domain for activity(Reference Bodine, Latres and Baumhueter6, Reference McElhinny, Kakinuma and Sorimachi7). There is growing evidence suggesting that long-chain EPA (C20 : 5n-3) can inhibit NF-κB activation by preventing the degradation of IκBα, further decreasing MuRF1 gene expression in skeletal muscle(Reference Castillero, Martín and López-Menduiña8, Reference Hommelberg, Plat and Langen9). However, the molecular mechanisms affected by n-3 PUFA, which leads to the inhibition of NF-κB activation, remain unclear.
Remarkably, activation of the transcription factor PPARγ in the skeletal muscle can inhibit NF-κB activity(Reference Gao, He and Peng10, Reference Remels, Langen and Gosker11). Previous studies have demonstrated that under normal physiological conditions, the dietary consumption of n-3 PUFA might activate PPARγ in the skeletal muscle of growing-finishing pigs(Reference Huang, Zhan and Luo12, Reference Zhan, Huang and Luo13). Aas et al. (Reference Aas, Rokling-Andersen and Kase14) also found that incubation of human skeletal muscle cells with 600 μm-EPA for 24 h increased PPARγ expression. Therefore, we hypothesised that under normal physiological conditions, n-3 PUFA might activate PPARγ, which may in turn inhibit the IκBα/NF-κB/MuRF1 signalling pathway in the skeletal muscle.
In the present study, C2C12 myotubes were treated with either α-linolenic acid (ALA; C18 : 3n-3) or EPA (C20 : 5n-3) for 24 h. Meanwhile, knockdown of PPARγ in C2C12 myotubes was achieved by RNA interference (RNAi). C2C12 myotubes with PPARγ knockdown were also treated with either ALA or EPA for 24 h. The actions of ALA or EPA were compared with those of a fatty acid-free control (containing bovine serum albumin (BSA)). The aim of the present study was to investigate the mechanism by which n-3 PUFA regulates the IκBα/NF-κB/MuRF1 pathway in C2C12 myotubes.
Materials and methods
Materials
Cell culture media and supplements were obtained from Invitrogen (Carlsbad, CA, USA). Reagents for complementary DNA synthesis and the LightCycler® system were obtained from Roche Applied Science (Mannheim, Germany). ALA (C18 : 3n-3), EPA (C20 : 5n-3), essentially fatty acid-free BSA, monoclonal anti-phospho-IκBα (Ser32) antibody and anti-IκBα antibody were obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA). Antibodies against NF-κB p65 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Finally, [γ-32P]ATP was obtained from Hartmann (Braunschweig, Germany).
Cell culture
Mouse C2C12 myoblasts (American Type Culture Collection, Manassas, VA, USA) were maintained in Dulbecco's modified Eagle's medium supplemented with 10 % fetal bovine serum, penicillin (50 units/ml) and streptomycin (50 μg/ml). When cells reached confluence, the medium was transferred to the differentiation medium containing Dulbecco's modified Eagle's medium and 2 % horse serum, which was changed every other day. After four additional days, the differentiated C2C12 cells had fused into myotubes.
Transfection of Stealth™ RNA interference for PPARγ knockdown in C2C12 myotubes
Transfection of Stealth™ RNAi for PPARγ knockdown in C2C12 myotubes was performed according to Kim et al. (Reference Kim, Cho and Jung15). The PPARγ Stealth™ Select RNAi oligonucleotide (target accession numbers NM138712.1, M015869.2, NM138711.1 and NM005037.3) was synthesised by Invitrogen (Carlsbad, CA, USA). The Stealth™ RNAi negative control duplex (Invitrogen Corporation, Carlsbad, CA, USA) was used as a control oligonucleotide. Transfection efficiency was monitored using a fluorescent oligonucleotide (BLOCK-iT fluorescent oligonucleotide; Invitrogen, Carlsbad, CA, USA) and was estimated to be 40 % in C2C12 cells. The Stealth™ RNAi molecules were then transfected into C2C12 myotubes using LipofectAMINE 2000 following Invitrogen's protocols. A final concentration of 50 nm of the PPARγ Stealth™ Select RNAi oligonucleotide was selected for C2C12 myotubes, and the Stealth™ RNAi oligonucleotides were transfected into the cells 48 h before treatment with fatty acids. The ability of the Stealth™ RNAi oligonucleotide to knock down PPARγ expression was analysed by Western blot and real-time quantitative PCR on whole-cell extracts.
NEFA treatment
Lipid-containing media were prepared by the conjugation of 150, 300 or 600 μm-NEFA (ALA or EPA) with NEFA-free BSA by a method modified from that described by Chavez et al. (Reference Chavez, Knotts and Wang16). Briefly, NEFA were dissolved in ethanol and diluted 1:100 in Dulbecco's modified Eagle's medium containing 300 μm-fatty acid-free BSA. Myotubes were then incubated for 24 h in serum-free Dulbecco's modified Eagle's medium containing 300 μm-BSA either in the presence (NEFA-treated cells) or in the absence (control cells) of NEFA. After the incubation, RNA was extracted from the myotubes as described in the following section.
RNA isolation
Total RNA was extracted using the TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer's specifications. The RNA samples were quantified spectrophotometrically at 260 and 280 nm. The ratio of light absorbance at 260 nm to that at 280 nm was between 1·8 and 2·0, indicating that they were pure and clean. The quality of RNA was also checked by 1·0 % agarose gel electrophoresis and staining with ethidium bromide (1 μg/ml).
Reverse transcription-PCR and real-time quantitative PCR analysis
Reverse transcription (20 μl) of total RNA (1 μg) was performed using an avian myeloblastosis virus RT with a first-strand complementary DNA synthesis kit for reverse transcription-PCR. Aliquots (2 μl) of the reverse transcription reactions were then submitted in duplicate to online quantitative PCR with the LightCycler® 480 Real-Time PCR system (Roche Applied Science) with SYBR green using the FastStart DNA-Master SYBR Green I kit (Roche Applied Science). Initial real-time amplifications were examined by agarose gel electrophoresis followed by ethidium bromide staining to verify that the primer pairs amplified a single product of the predicted size. Subsequent aliquots of the PCR were checked by melting curve analysis as provided by the LightCycler® 480 Real-Time PCR System (Roche Applied Science). Primer sequences and optimal PCR annealing temperatures (t a) are listed in Table 1. Specific primers were synthesised commercially (Shanghai Sangon Biological Engineering Technology and Services Company, Limited, Shanghai, China). The PCR were performed in a volume of 20 μl containing 2 μl of FastStart DNA-Master SYBR Green I kit, 3 mm-MgCl2 and primers at a concentration of 1 μm each. The instrument settings were denaturing at 95°C for 10 min, forty-five cycles of denaturing at 95°C for 30 s, annealing at 59°C for 30 s and elongating at 72°C for 8 min for PPARγ, MuRF1 and β-actin. Quantification was performed by online monitoring for identification of the exact time point at which the logarithmic linear phase was distinguishable from the background. Serially diluted samples obtained by PCR with the above-mentioned primers from human myotubes were used as external standards in each run. The cycle numbers for the logarithmic linear phase were plotted against the logarithm of the concentration of the template DNA, and the concentration of complementary DNA in the different samples was calculated with the LightCycler 5.32 software package (LC-Run version 5.32; Roche Applied Science).
Table 1 Oligonucleotide PCR primers
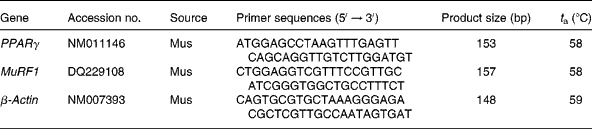
t a, Optimal PCR annealing temperature; MuRF1, muscle RING finger 1.
Isolation of nuclear extracts
Nuclear extracts were isolated according to Andrews et al. (Reference Andrews and Faller17). Cells were scraped into 1·5 ml of cold PBS, pelleted for 10 s and then resuspended in 400 μl of cold buffer A (10 mm-HEPES–KOH, pH 7·9 at 4°C, 1·5 mm-MgCl2, 10 mm-KCl, 0·5 mm-dithiothreitol, 0·2 mm-phenylmethylsulphonyl fluoride, 5 μg/ml aprotinin and 2 μg/ml leupeptin) by flicking the tube. Cells were allowed to swell on ice for 10 min and were then vortexed for 10 s. Samples were then centrifuged for 10 s, and the supernatant fraction was discarded. Pellets were resuspended in 50 μl of cold buffer C (20 mm-HEPES–KOH, pH 7·9 at 4°C, 25 % glycerol, 420 mm-NaCl, 1·5 mm-MgCl2, 0·2 mm-EDTA, 0·5 mm-dithiothreitol, 0·2 mm-phenylmethylsulphonyl fluoride, 5 μg/ml aprotinin and 2 μg/ml leupeptin) and incubated on ice for 20 min for high-salt extraction. Cellular debris was removed by centrifugation for 2 min at 4°C, and the supernatant fraction (containing DNA-binding proteins) was stored at − 80°C. The nuclear extract concentration was determined by the Bradford method.
Electrophoretic mobility shift assay
The transcription factor consensus oligonucleotides for the NF-κB-responsive element (5′-AGT TGA GGG GAC TTT CCC AGG C-3′) and the activator protein-1-responsive element (5′-CGC TTG ATG AGT CAG CCG GAA-3′) were purchased from Santa Cruz Biotechnology, Inc. The probes were labelled with [γ-32P]-ATP using T4 polynucleotide kinase (Boehringer-Mannheim, Lewes, UK) and purified on Sephadex G-25 spin chromatography columns (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, UK). For the binding reactions, the nuclear extract (1 μg of protein) was incubated in a 20 μl volume containing electrophoretic mobility shift assay-binding buffer (20 mm-HEPES, pH 7·5, 50 mm-NaCl, 1 mm-EDTA, 1 mm-dithiothreitol, 0·05 % NP-40 and 10 % glycerol), 0·25 ng [γ-32P]-labelled probe, BSA (1 mg/ml; Cell Signaling Technology, Inc.) and poly d(I-C) (100 ng/ml) for 20 min at room temperature. The specificity of the binding reaction was determined by co-incubating duplicate samples with either a 100-fold molar excess of an unlabelled oligonucleotide probe or an anti-NF-κB antibody (anti-p65; Santa Cruz Biotechnology, Inc.). Protein–nucleic acid complexes were resolved using a non-denaturing polyacrylamide gel consisting of 6 % acrylamide run in 5 mm-Tris (pH 8·3) and 38 mm-glycine for 2 h at 200 V. Gels were then transferred to Whatman 3M paper (Whatman, Inc., Clifton, NJ, USA), dried under a vacuum at 80°C for 1 h and exposed to a photographic film at − 70°C with an intensifying screen.
Immunoblotting
To obtain total proteins, C2C12 myotubes were homogenised in cold lysis buffer (5 mm-Tris–HCl, pH 7·4, 1 mm-EDTA, 0·1 mm-phenylmethylsulphonyl fluoride, 1 mm-sodium orthovanadate and 5·4 μg/ml aprotinin). The homogenate was then centrifuged at 10 000 g for 30 min at 4°C. For obtaining total membranes from C2C12 myotubes, cells were collected into 10 ml of ice-cold HES buffer (250 mm-sucrose, 1 m-EDTA, 1 m-phenylmethylsulphonyl fluoride, 1 μm-pepstatin, 1 μm-aprotinin, 1 μm-leupeptin and 20 m-HEPES, pH 7·4) and subsequently homogenised at 4°C. After centrifugation at 1000 g for 3 min at 4°C to remove large cell debris and unbroken cells, the supernatant was centrifuged at 245 000 g for 90 min at 4°C to yield a pellet of total cellular membranes. Proteins (30 μg) were separated by SDS-PAGE on 10 % separation gels and transferred to Immobilon polyvinylidene diflouride membranes (Millipore, Bedford, MA, USA). Western blot analysis was performed using antibodies against phospho-IκBα (Ser32), total IκBα and PPARγ (Santa Cruz Biotechnology, Inc.). Detection was achieved using the EZ-ECL chemiluminescence detection kit (Biological Industries, Beit Haemek Limited, Beit Haemek, Israel). The equal loading of proteins was assessed by red phenol staining. The size of the detected proteins was estimated using protein molecular mass standards (Invitrogen, Barcelona, Spain).
Statistical analysis
Data are presented as means with their standard errors. Differences between group means were determined by a one-way ANOVA using the computer program GraphPad Instat (version 2.03; GraphPad Software, Inc., San Diego, CA, USA). When significant variations were found, the Tukey–Kramer multiple comparison test was performed. Differences were considered significant at P < 0·05.
Results
Effect of 24 h treatment with 150, 300 or 600 μm-n-3 PUFA on the levels of phosphorylated inhibitor of κBα and total inhibitor of κBα in C2C12 myotubes
In the present study, two long-chain n-3 PUFA were chosen: ALA (a C18 : 3n-3 PUFA) and EPA (a C20 : 5n-3 PUFA). In addition, BSA was used as the fatty acid-free control. After treatment with 150, 300 or 600 μm-ALA and -EPA for 24 h, the levels of phosphorylated IκBα (p-IκBα) and total IκBα in C2C12 myotubes were measured by Western blot. Compared with the BSA control, a 24 h incubation of C2C12 myotubes with 150 μm (Fig. 1(a)) and 300 μm (Fig. 1(b)) ALA and EPA, respectively, did not affect the levels of p-IκBα and total IκBα.

Fig. 1 The effect of n-3 PUFA on the levels of phosphorylated inhibitor of κBα (p-IκBα) and total IκBα in C2C12 myotubes. C2C12 myotubes were incubated (24 h) with 150 μm (a), 300 μm (b) or 600 μm (c) of α-linolenic acid (ALA) or EPA. Bovine serum albumin (BSA) was used as the fatty acid-free control. Protein extracts from C2C12 myotubes were assayed by Western blot analysis by p-IκBα, total IκBα and β-actin. The band on the Western blot represented a protein with a molecular mass of approximately 37 kDa as determined by the molecular mass markers included in the experiment. The figure shows a representative blot of an experiment, reproduced independently at least three times.
The effect of 600 μm-ALA and -EPA on the IκBα protein levels in C2C12 myotubes is presented in Fig. 1(c). As expected, EPA (600 μm, 24 h) decreased IκBα phosphorylation and caused an approximately 86 % increase in total IκBα protein levels (P < 0·01). However, 600 μm-ALA did not affect the levels of p-IκBα or total IκBα (P>0·05). Taken together, these data suggested that 600 μm-EPA, but not ALA, prevented the degradation of IκBα by decreasing the phosphorylation of IκBα, increasing the total IκBα protein levels in C2C12 myotubes.
Effect of 24 h treatment with 600 μm-n-3 PUFA on the NF-κB-binding activity in C2C12 myotubes
To test whether incubation of C2C12 cells with 600 μm-ALA or -EPA for 24 h led to an effect on NF-κB activity, we performed electrophoretic mobility shift assay studies (Fig. 2). Compared with BSA, incubation of C2C12 myotubes with 600 μm-EPA for 24 h decreased the NF-κB DNA-binding activity in C2C12 myotubes (P < 0·01). Similar to the effect on the IκBα protein levels, 600 μm-ALA had no effect after 24 h on the NF-κB DNA-binding activity in C2C12 myotubes (P>0·05).

Fig. 2 NF-κB DNA-binding activity via the electrophoretic mobility shift assay (EMSA). C2C12 myotubes were incubated with 600 μm-α-linolenic acid (ALA) or 600 μm-EPA for 24 h. Bovine serum albumin (BSA) was used as the fatty acid-free control. Total nuclear protein was subsequently isolated and analysed by the EMSA for NF-κB DNA-binding activity using a 32P-labelled double-stranded oligonucleotide for the NF-κB. An additional non-labelled probe was added into the competition assay (cold). Data are representative of three independent experiments.
Effect of 24 h treatment with 600 μm-n-3 PUFA on the gene expression of muscle RING finger 1 and PPARγ in C2C12 myotubes
Because NF-κB is translocated to the cell nucleus upon activation, leading to the stimulation of MuRF1 gene expression(Reference Cai, Frantz and Tawa5), we next investigated whether incubation of C2C12 cells with 600 μm-ALA or -EPA for 24 h affected MuRF1 mRNA expression (Fig. 3(b)). C2C12 myotubes incubated in the presence of 600 μm-EPA for 24 h caused a 3·38-fold reduction in the levels of MuRF1 mRNA (P < 0·01). However, compared with BSA, 600 μm-ALA did not affect MuRF1 mRNA expression (P>0·05).

Fig. 3 Effects of n-3 PUFA on the PPARγ and muscle RING finger 1 (MuRF1) gene expression. C2C12 myotubes were incubated with 600 μm-α-linolenic acid (ALA, ■) or 600 μm-EPA (![]() ) for 24 h. Bovine serum albumin (BSA, □) was used as the fatty acid-free control. The PPARγ mRNA and MuRF1 mRNA levels were determined using real-time PCR analysis, and the relative abundance of mRNA was calculated after normalisation to β-actin. PPARγ (a) and MuRF1 (b). Data are expressed as the mean values and standard deviations of three different experiments. ** Mean values were significantly different from that of the control group (P < 0·01).
) for 24 h. Bovine serum albumin (BSA, □) was used as the fatty acid-free control. The PPARγ mRNA and MuRF1 mRNA levels were determined using real-time PCR analysis, and the relative abundance of mRNA was calculated after normalisation to β-actin. PPARγ (a) and MuRF1 (b). Data are expressed as the mean values and standard deviations of three different experiments. ** Mean values were significantly different from that of the control group (P < 0·01).
PPARγ gene expression data are presented in Fig. 3(a). Incubation of C2C12 myotubes in the presence of 600 μm-EPA for 24 h resulted in a 2·3-fold induction of PPARγ expression (P < 0·01), whereas 24 h incubation with 600 μm-ALA had no effect (P>0·05).
Effect of 24 h treatment with 600 μm-n-3 PUFA on the inhibitor of κBα/NF-κB/muscle RING finger 1 pathway in C2C12 myotubes with PPARγ knockdown
To confirm that the inhibition of the IκBα/NF-κB/MuRF1 pathway by n-3 PUFA is mediated via activation of PPARγ mRNA expression, we examined the effect of n-3 PUFA on the IκBα/NF-κB/MuRF1 pathway in C2C12 myotubes with PPARγ knockdown. As such, the C2C12 myotubes transfected with either the negative control Stealth™ RNAi oligonucleotide or the PPARγ Stealth™ RNAi oligonucleotide were incubated for 48 h. Transfection of Stealth™ RNAi for PPARγ knockdown in C2C12 myotubes was found to significantly decrease PPARγ protein expression (Fig. 4(a) and (b)) and PPARγ mRNA expression (Fig. 4(c)) to approximately 50 and 60 % of their normal levels (P < 0·01), respectively. Negative control Stealth™ RNAi treatment had no influence on PPARγ mRNA and protein expression (P>0·05).

Fig. 4 Transfection of Stealth™ RNA interference (RNAi) for PPARγ knockdown in C2C12 myotubes. The C2C12 myotubes transfected with either the negative control Stealth™ RNAi oligonucleotide or the PPARγ Stealth™ RNAi oligonucleotide were incubated for 48 h. The Stealth™ RNAi negative control duplexes with similar G/C content (Invitrogen, Carlsbad, CA, USA) were used as negative controls. Protein extracts from C2C12 myotubes were assayed by Western blot analysis for PPARγ (a). The band on the Western blot represented a protein with a molecular mass of approximately 55 kDa, as determined by the molecular mass markers included in the experiment. PPARγ protein expression was determined by Western blot, and the relative abundance of protein was calculated after normalisation to β-actin (b). PPARγ mRNA was determined using real-time PCR analysis, and the relative abundance of mRNA was calculated after normalisation to β-actin (c). Data are expressed as the mean values and standard deviations of three independent experiments. ** Mean values were significantly different from that of the untransfected control group (P < 0·01). □, Untransfected control; ■, negative control Stealth™ RNAi-transfected cell; ![]() , PPARγ Stealth™ RNAi-transfected cell.
, PPARγ Stealth™ RNAi-transfected cell.
Compared with the Stealth™ RNAi-transfected control, 600 μm-EPA, but not ALA, prevented the degradation of IκBα by decreasing the phosphorylation of IκBα (P < 0·01), thus increasing the total IκBα protein levels (Fig. 5(a)) in C2C12 myotubes transfected with the negative control Stealth™ RNAi oligonucleotide (P < 0·01). It was further observed that 600 μm-EPA decreased NF-κB DNA-binding activity (Fig. 5(b)) and inhibited MuRF1 mRNA expression (Fig. 6) in C2C12 myotubes transfected with the negative control Stealth™ RNAi oligonucleotide (P < 0·01). However, in C2C12 myotubes transfected with the PPARγ Stealth™ RNAi oligonucleotide, treatment with 600 μm-ALA or -EPA for 24 h did not affect the levels of p-IκBα or total IκBα (Fig. 5(a)), NF-κB DNA-binding activity (Fig. 5(b)) or MuRF1 mRNA expression (Fig. 6) compared with the PPARγ Stealth™ RNAi-transfected control (P>0·05).

Fig. 5 The effect of n-3 PUFA on the inhibitor of κBα (IκBα)/NF-κB complex in C2C12 myotubes transfected with the PPARγ Stealth™ RNA interference (RNAi) oligonucleotide. After the C2C12 myotubes were transfected with either the negative control Stealth™ RNAi oligonucleotide or the PPARγ Stealth™ RNA interference (RNAi) oligonucleotide for 48 h, C2C12 myotubes were incubated with 600 μm-α-linolenic acid (ALA) or 600 μm-EPA for 24 h. Bovine serum albumin (BSA) was used as the fatty acid-free control. Protein extracts from C2C12 myotubes were assayed by Western blot analysis for phosphorylated IκBα, total IκBα and β-actin (a). The band on the Western blot represented a protein with a molecular mass of approximately 37 kDa, as determined by the molecular mass markers included in the experiment. Total nuclear protein was subsequently isolated and analysed by the electrophoretic mobility shift assay for NF-κB DNA-binding activity using a 32P-labelled double-stranded oligonucleotide for NF-κB (b). An additional non-labelled probe was added to the competition assay (cold). Data are representative of three independent experiments.

Fig. 6 The effect of n-3 PUFA on the muscle RING finger 1 (MuRF1) gene expression in C2C12 myotubes transfected with the PPARγ Stealth™ RNA interference (RNAi) oligonucleotide. After the transfection of C2C12 myotubes with either the negative control Stealth™ RNAi oligonucleotide or the PPARγ Stealth™ RNAi oligonucleotide for 48 h, C2C12 myotubes were incubated with 600 μm-α-linolenic acid (ALA, ■) or 600 μm-EPA (![]() ) for 24 h. Bovine serum albumin (BSA, □) was used as the fatty acid-free control. MuRF1 mRNA was determined using real-time PCR analysis, and the relative abundance of mRNA was calculated after normalisation to β-actin. Data are expressed as the mean values and standard deviations of three independent experiments. ** Mean values were significantly different from that of the negative Stealth™ RNAi-transfected control group (P < 0·01).
) for 24 h. Bovine serum albumin (BSA, □) was used as the fatty acid-free control. MuRF1 mRNA was determined using real-time PCR analysis, and the relative abundance of mRNA was calculated after normalisation to β-actin. Data are expressed as the mean values and standard deviations of three independent experiments. ** Mean values were significantly different from that of the negative Stealth™ RNAi-transfected control group (P < 0·01).
Discussion
The key to NF-κB regulation is the IκBα protein, which is retained in the cytoplasm. Phosphorylation of IκBα by IκB kinases triggers its polyubiquitinylation and degradation, thereby releasing NF-κB, which can then translocate to the nucleus. In the present study, C2C12 myotubes incubated in the presence of 150, 300 or 600 μm-ALA for 24 h did not have different levels of p-IκBα or total IκBα compared with the fatty acid-free BSA control, suggesting that ALA was not able to regulate NF-κB activity by preventing phosphorylation of IκBα and decreasing total IκBα degradation in C2C12 myotubes. Interestingly, the addition of 600 μm-EPA (C20 : 5n-3) to the cells decreased IκBα phosphorylation and caused an approximate 86 % increase in total IκBα protein levels. It was further observed that 600 μm-EPA inhibited NF-κB activation in C2C12 myotubes. These results are in general agreement with other studies showing that EPA inhibited NF-κB activation by preventing IκBα phosphorylation, thus further reducing the degradation of the inhibitory IκBα protein(Reference Whitehouse and Tisdale18, Reference Babcock, Helton and Espat19).
In the present study, there was no effect on the levels of p-IκBα or total IκBα following a chronic exposure to 150 or 300 μm-EPA, suggesting that the effect of EPA on the levels of p-IκBα and total IκBα may be dependent on the concentration of the fatty acid. Remarkably, Lo et al. (Reference Lo, Chiu and Fu20) reported that proteolysis-inducing factor, isolated from a cachexia-inducing murine tumour, activated NF-κB by inducing the degradation of IκBα in C2C12 myotubes, and that 50 μm-EPA effectively attenuated the proteolysis-inducing factor-induced IκBα/NF-κB pathway under various pathophysiological conditions. Therefore, the present study provides evidence that the effect of EPA on the IκBα/NF-κB pathway requires higher concentrations of fatty acids under normal physiological conditions than under pathophysiological conditions.
MuRF1 is a 40 kDa protein that contains a RING domain at its amino-terminal end as well as two coiled-coil domains in its central region, and is found in the nuclei of muscle cells(Reference Centner, Yano and Kimura21, Reference Spencer and Christensen22). MuRF1 has been demonstrated to have ubiquitin-ligase activity that requires the presence of the RING domain for normal activity(Reference Bodine, Latres and Baumhueter6). Previous studies revealed that the NF-κB was translocated to the cell nucleus upon activation and led to the stimulation of MuRF1 gene expression(Reference Cai, Frantz and Tawa5). In the present study, incubation of C2C12 myotubes in the presence of 600 μm-EPA for 24 h caused a 3·38-fold induction in the level of MuRF1 mRNA, whereas a 24 h incubation period with 600 μm-ALA did not affect MuRF1 mRNA expression. These results revealed that EPA, but not ALA, inhibited NF-κB activation by preventing the degradation of IκBα and further decreasing the MuRF1 gene expression in C2C12 myotubes.
We next investigated the effect of incubation of C2C12 myotubes with 600 μm-ALA or -EPA for 24 h on PPARγ gene expression. PPARγ is a member of the nuclear receptor superfamily of PPAR, and its main function was originally thought to regulate adipocyte differentiation(Reference Pégorier, Le May and Girard23, Reference Hammad, de Heer and Soullié24). Recently, growing evidence points to its implication in the regulation of the immune response, particularly in inflammation control(Reference Sokołowska, Kowalski and Pawliczak25). The strongest PPARγ expression has been observed in adipose tissues and in the spleen, with a lower expression in the kidney, large intestine, heart, lung, small intestine and skeletal muscle(Reference Grindflek, Sundvold and Klungland26).
In the present study, it was observed that C2C12 myotubes incubated in the presence of EPA (600 μm, 24 h) resulted in a 1·47-fold induction of PPARγ expression compared with the BSA control, which is in line with a previous report by Aas et al. (Reference Aas, Rokling-Andersen and Kase14), who found a 2·3-fold increase in PPARγ expression after exposure of human skeletal muscle cells to EPA (600 μm, 24 h). It is noteworthy that C2C12 myotubes incubated in the presence of ALA (600 μm, 24 h) had no change in PPARγ mRNA levels. Chambrier et al. (Reference Chambrier, Bastard and Rieusset27) also found that EPA (50 μm, 6 h) induced PPARγ mRNA levels in human adipocytes, but that ALA (50 μm, 6 h), a precursor of EPA, did not change PPARγ mRNA levels. Furthermore, they observed that it was likely that the metabolism of ALA was not efficient enough to lead to an effective concentration of EPA in human adipocytes(Reference Chambrier, Bastard and Rieusset27). Taken together, these results demonstrated that long-chain EPA could induce PPARγ mRNA expression in skeletal muscle cells, while ALA, with a shorter chain length, was not able to affect PPARγ mRNA expression.
Additionally, the present results revealed that EPA, but not ALA, was effective at inhibiting the IκBα/NF-κB/MuRF1 pathway in C2C12 myotubes. However, the reason why ALA could not regulate the IκBα/NF-κB/MuRF1 pathway in C2C12 myotubes is poorly documented. Interestingly, we discovered that EPA could induce PPARγ mRNA expression in skeletal muscle cells, while ALA was not able to affect PPARγ mRNA expression. Therefore, the reason why ALA could not regulate the IκBα/NF-κB/MuRF1 pathway may have been that ALA was not able to affect PPARγ mRNA expression in C2C12 myotubes.
To investigate whether EPA inhibited the IκBα/NF-κB/MuRF1 pathway via activation of PPARγ mRNA expression, PPARγ knockdown by RNAi was used to decrease PPARγ mRNA and protein expression to approximately 50 and 60 %, respectively, of their normal levels in C2C12 myotubes. Interestingly, it was observed that treatment with 600 μm-EPA for 24 h did not affect the levels of p-IκBα, total IκBα, NF-κB DNA-binding activity or MuRF1 mRNA expression in C2C12 myotubes with PPARγ knockdown. These results demonstrated that PPARγ knockdown by RNAi abolished the suppressive effects of EPA on the IκBα/NF-κB/MuRF1 pathway in C2C12 myotubes, supporting the claim that the effects of EPA are mediated via PPARγ activation. Taken together, these results revealed that EPA, but not ALA, activated PPARγ, which may in turn have inhibited the IκBα/NF-κB/MuRF1 signalling pathway in C2C12 myotubes.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (nos 30972107 and 30871779), the Major Science & Technology Industrialization Projects Program in the city of Wuhan (no. 200720112026), the National High Technology R&D Program of China (no. 2006AA10Z140) and the International Foundation for Science (no. B/4909-1). All authors contributed to the preparation of the paper and agreed with the content of the submitted manuscript. F. H., J. P. and S. J. designed the research; F. H., H. W. and H. L. performed the research; F. H., J. P. and S. J. analysed the data and wrote the paper. All authors declare that there are no conflicts of interest.











